Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
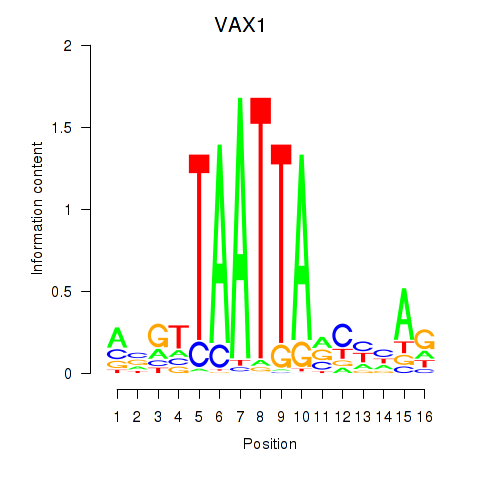
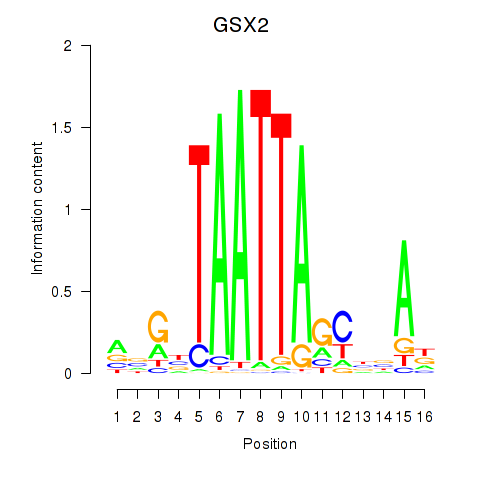
Results for VAX1_GSX2
Z-value: 0.48
Transcription factors associated with VAX1_GSX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
VAX1
|
ENSG00000148704.8 | ventral anterior homeobox 1 |
|
GSX2
|
ENSG00000180613.6 | GS homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| VAX1 | hg19_v2_chr10_-_118897567_118897600 | -0.57 | 4.3e-01 | Click! |
Activity profile of VAX1_GSX2 motif
Sorted Z-values of VAX1_GSX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_99716914 | 0.61 |
ENST00000431404.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr11_+_115498761 | 0.29 |
ENST00000424313.2
|
AP000997.1
|
AP000997.1 |
| chr2_-_99871570 | 0.25 |
ENST00000333017.2
ENST00000409679.1 ENST00000423306.1 |
LYG2
|
lysozyme G-like 2 |
| chr5_+_53686658 | 0.19 |
ENST00000512618.1
|
LINC01033
|
long intergenic non-protein coding RNA 1033 |
| chr9_+_131062367 | 0.18 |
ENST00000601297.1
|
AL359091.2
|
CDNA: FLJ21673 fis, clone COL09042; HCG2036511; Uncharacterized protein |
| chr17_+_73539339 | 0.18 |
ENST00000581713.1
|
LLGL2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr5_-_139726181 | 0.18 |
ENST00000507104.1
ENST00000230990.6 |
HBEGF
|
heparin-binding EGF-like growth factor |
| chr12_+_78359999 | 0.18 |
ENST00000550503.1
|
NAV3
|
neuron navigator 3 |
| chr11_+_29181503 | 0.17 |
ENST00000530960.1
|
RP11-466I1.1
|
RP11-466I1.1 |
| chr17_+_28443819 | 0.16 |
ENST00000479218.2
|
NSRP1
|
nuclear speckle splicing regulatory protein 1 |
| chr4_+_144354644 | 0.16 |
ENST00000512843.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr6_+_160693591 | 0.15 |
ENST00000419196.1
|
RP1-276N6.2
|
RP1-276N6.2 |
| chr19_+_48949087 | 0.15 |
ENST00000598711.1
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chr15_-_36544450 | 0.15 |
ENST00000561394.1
|
RP11-184D12.1
|
RP11-184D12.1 |
| chr4_+_70894130 | 0.14 |
ENST00000526767.1
ENST00000530128.1 ENST00000381057.3 |
HTN3
|
histatin 3 |
| chr2_+_90198535 | 0.14 |
ENST00000390276.2
|
IGKV1D-12
|
immunoglobulin kappa variable 1D-12 |
| chr4_-_112993808 | 0.14 |
ENST00000511219.1
|
RP11-269F21.3
|
RP11-269F21.3 |
| chr17_-_57229155 | 0.13 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr13_+_78315295 | 0.13 |
ENST00000351546.3
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr4_-_89442940 | 0.13 |
ENST00000527353.1
|
PIGY
|
phosphatidylinositol glycan anchor biosynthesis, class Y |
| chr12_-_57039739 | 0.12 |
ENST00000552959.1
ENST00000551020.1 ENST00000553007.2 ENST00000552919.1 ENST00000552104.1 ENST00000262030.3 |
ATP5B
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr17_+_74846230 | 0.12 |
ENST00000592919.1
|
LINC00868
|
long intergenic non-protein coding RNA 868 |
| chr1_+_192778161 | 0.12 |
ENST00000235382.5
|
RGS2
|
regulator of G-protein signaling 2, 24kDa |
| chr19_+_48949030 | 0.12 |
ENST00000253237.5
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chr1_-_201140673 | 0.11 |
ENST00000367333.2
|
TMEM9
|
transmembrane protein 9 |
| chr7_+_99202003 | 0.11 |
ENST00000609449.1
|
GS1-259H13.2
|
GS1-259H13.2 |
| chr5_+_135394840 | 0.11 |
ENST00000503087.1
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chr11_+_112041253 | 0.10 |
ENST00000532612.1
|
AP002884.3
|
AP002884.3 |
| chr1_+_54569968 | 0.10 |
ENST00000391366.1
|
AL161915.1
|
Uncharacterized protein |
| chr10_-_128110441 | 0.10 |
ENST00000456514.1
|
LINC00601
|
long intergenic non-protein coding RNA 601 |
| chr11_-_124981475 | 0.10 |
ENST00000532156.1
ENST00000532407.1 ENST00000279968.4 ENST00000527766.1 ENST00000529583.1 ENST00000524373.1 ENST00000527271.1 ENST00000526175.1 ENST00000529609.1 ENST00000533273.1 ENST00000531909.1 ENST00000529530.1 |
TMEM218
|
transmembrane protein 218 |
| chr12_-_10282742 | 0.09 |
ENST00000298523.5
ENST00000396484.2 ENST00000310002.4 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr6_+_155538093 | 0.09 |
ENST00000462408.2
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr17_-_46690839 | 0.09 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr14_+_32798547 | 0.09 |
ENST00000557354.1
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr10_-_28571015 | 0.09 |
ENST00000375719.3
ENST00000375732.1 |
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr5_+_174151536 | 0.09 |
ENST00000239243.6
ENST00000507785.1 |
MSX2
|
msh homeobox 2 |
| chr15_+_63188009 | 0.08 |
ENST00000557900.1
|
RP11-1069G10.2
|
RP11-1069G10.2 |
| chr7_-_81399438 | 0.08 |
ENST00000222390.5
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr14_-_71609965 | 0.08 |
ENST00000602957.1
|
RP6-91H8.3
|
RP6-91H8.3 |
| chr6_-_116833500 | 0.08 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr14_+_67831576 | 0.08 |
ENST00000555876.1
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa |
| chr8_+_107738343 | 0.08 |
ENST00000521592.1
|
OXR1
|
oxidation resistance 1 |
| chr2_-_101925055 | 0.08 |
ENST00000295317.3
|
RNF149
|
ring finger protein 149 |
| chr1_-_36789755 | 0.08 |
ENST00000270824.1
|
EVA1B
|
eva-1 homolog B (C. elegans) |
| chr7_+_100860949 | 0.07 |
ENST00000305105.2
|
ZNHIT1
|
zinc finger, HIT-type containing 1 |
| chr19_-_58951496 | 0.07 |
ENST00000254166.3
|
ZNF132
|
zinc finger protein 132 |
| chr2_-_133429091 | 0.07 |
ENST00000345008.6
|
LYPD1
|
LY6/PLAUR domain containing 1 |
| chr17_-_72772462 | 0.07 |
ENST00000582870.1
ENST00000581136.1 ENST00000357814.3 ENST00000579218.1 ENST00000583476.1 ENST00000580301.1 ENST00000583757.1 ENST00000582524.1 |
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr19_+_50016610 | 0.07 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr13_+_36050881 | 0.07 |
ENST00000537702.1
|
NBEA
|
neurobeachin |
| chr6_+_26365443 | 0.06 |
ENST00000527422.1
ENST00000356386.2 ENST00000396934.3 ENST00000377708.2 ENST00000396948.1 ENST00000508906.2 |
BTN3A2
|
butyrophilin, subfamily 3, member A2 |
| chr20_+_43990576 | 0.06 |
ENST00000372727.1
ENST00000414310.1 |
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr12_+_113587558 | 0.06 |
ENST00000335621.6
|
CCDC42B
|
coiled-coil domain containing 42B |
| chr5_+_142286887 | 0.06 |
ENST00000451259.1
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr17_-_72772425 | 0.06 |
ENST00000578822.1
|
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr17_-_4938712 | 0.06 |
ENST00000254853.5
ENST00000424747.1 |
SLC52A1
|
solute carrier family 52 (riboflavin transporter), member 1 |
| chr6_+_130339710 | 0.06 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr5_-_96478466 | 0.06 |
ENST00000274382.4
|
LIX1
|
Lix1 homolog (chicken) |
| chr19_+_45417921 | 0.06 |
ENST00000252491.4
ENST00000592885.1 ENST00000589781.1 |
APOC1
|
apolipoprotein C-I |
| chrX_-_23926004 | 0.06 |
ENST00000379226.4
ENST00000379220.3 |
APOO
|
apolipoprotein O |
| chr1_-_207226313 | 0.06 |
ENST00000367084.1
|
YOD1
|
YOD1 deubiquitinase |
| chr12_-_57328187 | 0.06 |
ENST00000293502.1
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C, member 7 |
| chr2_-_198540719 | 0.06 |
ENST00000295049.4
|
RFTN2
|
raftlin family member 2 |
| chr3_-_180397256 | 0.06 |
ENST00000442201.2
|
CCDC39
|
coiled-coil domain containing 39 |
| chr18_+_3447572 | 0.06 |
ENST00000548489.2
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr8_-_41166953 | 0.06 |
ENST00000220772.3
|
SFRP1
|
secreted frizzled-related protein 1 |
| chr20_-_50722183 | 0.06 |
ENST00000371523.4
|
ZFP64
|
ZFP64 zinc finger protein |
| chr17_-_73901494 | 0.06 |
ENST00000309352.3
|
MRPL38
|
mitochondrial ribosomal protein L38 |
| chr1_+_36789335 | 0.06 |
ENST00000373137.2
|
RP11-268J15.5
|
RP11-268J15.5 |
| chr8_+_107738240 | 0.05 |
ENST00000449762.2
ENST00000297447.6 |
OXR1
|
oxidation resistance 1 |
| chr15_+_36871983 | 0.05 |
ENST00000437989.2
ENST00000569302.1 |
C15orf41
|
chromosome 15 open reading frame 41 |
| chrX_+_107288239 | 0.05 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr5_-_41794313 | 0.05 |
ENST00000512084.1
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chr4_+_80584903 | 0.05 |
ENST00000506460.1
|
RP11-452C8.1
|
RP11-452C8.1 |
| chr5_+_129083772 | 0.05 |
ENST00000564719.1
|
KIAA1024L
|
KIAA1024-like |
| chr16_-_66764119 | 0.05 |
ENST00000569320.1
|
DYNC1LI2
|
dynein, cytoplasmic 1, light intermediate chain 2 |
| chr12_-_112123524 | 0.05 |
ENST00000327551.6
|
BRAP
|
BRCA1 associated protein |
| chr15_+_62853562 | 0.05 |
ENST00000561311.1
|
TLN2
|
talin 2 |
| chr15_-_65903407 | 0.05 |
ENST00000395644.4
ENST00000567744.1 ENST00000568573.1 ENST00000562830.1 ENST00000569491.1 ENST00000561769.1 |
VWA9
|
von Willebrand factor A domain containing 9 |
| chr1_+_160370344 | 0.05 |
ENST00000368061.2
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr11_-_26593649 | 0.05 |
ENST00000455601.2
|
MUC15
|
mucin 15, cell surface associated |
| chr14_+_101359265 | 0.05 |
ENST00000599197.1
|
AL117190.3
|
Esophagus cancer-related gene-2 interaction susceptibility protein; Uncharacterized protein |
| chr11_-_795400 | 0.05 |
ENST00000526152.1
ENST00000456706.2 ENST00000528936.1 |
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr18_-_46895066 | 0.05 |
ENST00000583225.1
ENST00000584983.1 ENST00000583280.1 ENST00000581738.1 |
DYM
|
dymeclin |
| chr3_-_20053741 | 0.05 |
ENST00000389050.4
|
PP2D1
|
protein phosphatase 2C-like domain containing 1 |
| chr2_-_220264703 | 0.05 |
ENST00000519905.1
ENST00000523282.1 ENST00000434339.1 ENST00000457935.1 |
DNPEP
|
aspartyl aminopeptidase |
| chr19_+_11071546 | 0.05 |
ENST00000358026.2
|
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr10_-_92681033 | 0.05 |
ENST00000371697.3
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle) |
| chr12_-_10282836 | 0.05 |
ENST00000304084.8
ENST00000353231.5 ENST00000525605.1 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr14_-_54423529 | 0.05 |
ENST00000245451.4
ENST00000559087.1 |
BMP4
|
bone morphogenetic protein 4 |
| chr1_-_1709845 | 0.05 |
ENST00000341426.5
ENST00000344463.4 |
NADK
|
NAD kinase |
| chr10_+_5005598 | 0.05 |
ENST00000442997.1
|
AKR1C1
|
aldo-keto reductase family 1, member C1 |
| chr15_+_92006567 | 0.05 |
ENST00000554333.1
|
RP11-661P17.1
|
RP11-661P17.1 |
| chr1_-_202897724 | 0.05 |
ENST00000435533.3
ENST00000367258.1 |
KLHL12
|
kelch-like family member 12 |
| chr7_+_100770328 | 0.04 |
ENST00000223095.4
ENST00000445463.2 |
SERPINE1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
| chr11_+_92085262 | 0.04 |
ENST00000298047.6
ENST00000409404.2 ENST00000541502.1 |
FAT3
|
FAT atypical cadherin 3 |
| chr4_+_144312659 | 0.04 |
ENST00000509992.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr11_-_795286 | 0.04 |
ENST00000533385.1
ENST00000527723.1 |
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr11_-_795170 | 0.04 |
ENST00000481290.1
|
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr15_+_84115868 | 0.04 |
ENST00000427482.2
|
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr9_-_19149276 | 0.04 |
ENST00000434144.1
|
PLIN2
|
perilipin 2 |
| chrX_+_107288197 | 0.04 |
ENST00000415430.3
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr6_+_26440700 | 0.04 |
ENST00000494393.1
ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3
|
butyrophilin, subfamily 3, member A3 |
| chr6_+_26365387 | 0.04 |
ENST00000532865.1
ENST00000530653.1 ENST00000527417.1 |
BTN3A2
|
butyrophilin, subfamily 3, member A2 |
| chr17_+_12569472 | 0.04 |
ENST00000343344.4
|
MYOCD
|
myocardin |
| chr15_-_55562582 | 0.04 |
ENST00000396307.2
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr11_+_100862811 | 0.04 |
ENST00000303130.2
|
TMEM133
|
transmembrane protein 133 |
| chr11_-_65149422 | 0.04 |
ENST00000526432.1
ENST00000527174.1 |
SLC25A45
|
solute carrier family 25, member 45 |
| chr7_-_99717463 | 0.04 |
ENST00000437822.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr6_+_13272904 | 0.04 |
ENST00000379335.3
ENST00000379329.1 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr22_-_39636914 | 0.04 |
ENST00000381551.4
|
PDGFB
|
platelet-derived growth factor beta polypeptide |
| chr9_+_125133315 | 0.04 |
ENST00000223423.4
ENST00000362012.2 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr4_-_109541539 | 0.04 |
ENST00000509984.1
ENST00000507248.1 ENST00000506795.1 |
RPL34-AS1
|
RPL34 antisense RNA 1 (head to head) |
| chr19_+_17638059 | 0.04 |
ENST00000599164.1
ENST00000449408.2 ENST00000600871.1 ENST00000599124.1 |
FAM129C
|
family with sequence similarity 129, member C |
| chr8_-_55014018 | 0.04 |
ENST00000521352.1
|
LYPLA1
|
lysophospholipase I |
| chr3_+_152552685 | 0.04 |
ENST00000305097.3
|
P2RY1
|
purinergic receptor P2Y, G-protein coupled, 1 |
| chr16_+_31885079 | 0.04 |
ENST00000300870.10
ENST00000394846.3 |
ZNF267
|
zinc finger protein 267 |
| chr1_-_1710229 | 0.04 |
ENST00000341991.3
|
NADK
|
NAD kinase |
| chr19_+_21264980 | 0.04 |
ENST00000596053.1
ENST00000597086.1 ENST00000596143.1 ENST00000596367.1 ENST00000601416.1 |
ZNF714
|
zinc finger protein 714 |
| chr18_-_44181442 | 0.04 |
ENST00000398722.4
|
LOXHD1
|
lipoxygenase homology domains 1 |
| chr2_-_166930131 | 0.04 |
ENST00000303395.4
ENST00000409050.1 ENST00000423058.2 ENST00000375405.3 |
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit |
| chr11_+_46958248 | 0.04 |
ENST00000536126.1
ENST00000278460.7 ENST00000378618.2 ENST00000395460.2 ENST00000378615.3 ENST00000543718.1 |
C11orf49
|
chromosome 11 open reading frame 49 |
| chr2_+_44502597 | 0.04 |
ENST00000260649.6
ENST00000409387.1 |
SLC3A1
|
solute carrier family 3 (amino acid transporter heavy chain), member 1 |
| chr8_+_92261516 | 0.04 |
ENST00000276609.3
ENST00000309536.2 |
SLC26A7
|
solute carrier family 26 (anion exchanger), member 7 |
| chr9_-_132383055 | 0.03 |
ENST00000372478.4
|
C9orf50
|
chromosome 9 open reading frame 50 |
| chr4_-_39979576 | 0.03 |
ENST00000303538.8
ENST00000503396.1 |
PDS5A
|
PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae) |
| chr7_-_100860851 | 0.03 |
ENST00000223127.3
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr17_+_18086392 | 0.03 |
ENST00000541285.1
|
ALKBH5
|
alkB, alkylation repair homolog 5 (E. coli) |
| chr11_-_129062093 | 0.03 |
ENST00000310343.9
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr1_+_28261492 | 0.03 |
ENST00000373894.3
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr16_+_29832634 | 0.03 |
ENST00000565164.1
ENST00000570234.1 |
MVP
|
major vault protein |
| chr19_+_36632204 | 0.03 |
ENST00000592354.1
|
CAPNS1
|
calpain, small subunit 1 |
| chr10_-_135379132 | 0.03 |
ENST00000343131.5
|
SYCE1
|
synaptonemal complex central element protein 1 |
| chr11_+_24518723 | 0.03 |
ENST00000336930.6
ENST00000529015.1 ENST00000533227.1 |
LUZP2
|
leucine zipper protein 2 |
| chr15_-_55562479 | 0.03 |
ENST00000564609.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr16_-_31085514 | 0.03 |
ENST00000300849.4
|
ZNF668
|
zinc finger protein 668 |
| chr15_+_40697988 | 0.03 |
ENST00000487418.2
ENST00000479013.2 |
IVD
|
isovaleryl-CoA dehydrogenase |
| chr4_-_66536057 | 0.03 |
ENST00000273854.3
|
EPHA5
|
EPH receptor A5 |
| chrX_+_108779004 | 0.03 |
ENST00000218004.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr3_-_99569821 | 0.03 |
ENST00000487087.1
|
FILIP1L
|
filamin A interacting protein 1-like |
| chr13_-_46626820 | 0.03 |
ENST00000428921.1
|
ZC3H13
|
zinc finger CCCH-type containing 13 |
| chr1_+_214161854 | 0.03 |
ENST00000435016.1
|
PROX1
|
prospero homeobox 1 |
| chr3_+_51851612 | 0.03 |
ENST00000456080.1
|
IQCF3
|
IQ motif containing F3 |
| chr2_+_105050794 | 0.03 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr12_+_26348246 | 0.03 |
ENST00000422622.2
|
SSPN
|
sarcospan |
| chr19_+_19303008 | 0.03 |
ENST00000353145.1
ENST00000421262.3 ENST00000303088.4 ENST00000456252.3 ENST00000593273.1 |
RFXANK
|
regulatory factor X-associated ankyrin-containing protein |
| chr2_+_191221240 | 0.03 |
ENST00000409027.1
ENST00000458193.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr12_+_56862301 | 0.03 |
ENST00000338146.5
|
SPRYD4
|
SPRY domain containing 4 |
| chr3_+_149191723 | 0.03 |
ENST00000305354.4
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chrX_-_16887963 | 0.03 |
ENST00000380084.4
|
RBBP7
|
retinoblastoma binding protein 7 |
| chr19_+_21324827 | 0.03 |
ENST00000600692.1
ENST00000599296.1 ENST00000594425.1 ENST00000311048.7 |
ZNF431
|
zinc finger protein 431 |
| chr21_-_43346790 | 0.03 |
ENST00000329623.7
|
C2CD2
|
C2 calcium-dependent domain containing 2 |
| chr9_+_5450503 | 0.03 |
ENST00000381573.4
ENST00000381577.3 |
CD274
|
CD274 molecule |
| chr3_+_62304712 | 0.03 |
ENST00000494481.1
|
C3orf14
|
chromosome 3 open reading frame 14 |
| chr12_+_20963647 | 0.02 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr19_+_50180409 | 0.02 |
ENST00000391851.4
|
PRMT1
|
protein arginine methyltransferase 1 |
| chr17_+_72427477 | 0.02 |
ENST00000342648.5
ENST00000481232.1 |
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr12_+_7014064 | 0.02 |
ENST00000443597.2
|
LRRC23
|
leucine rich repeat containing 23 |
| chr2_+_32288725 | 0.02 |
ENST00000315285.3
|
SPAST
|
spastin |
| chr1_+_158978768 | 0.02 |
ENST00000447473.2
|
IFI16
|
interferon, gamma-inducible protein 16 |
| chr10_+_99205959 | 0.02 |
ENST00000352634.4
ENST00000353979.3 ENST00000370842.2 ENST00000345745.5 |
ZDHHC16
|
zinc finger, DHHC-type containing 16 |
| chr19_+_50180507 | 0.02 |
ENST00000454376.2
ENST00000524771.1 |
PRMT1
|
protein arginine methyltransferase 1 |
| chr7_+_37723420 | 0.02 |
ENST00000476620.1
|
EPDR1
|
ependymin related 1 |
| chr3_+_108541545 | 0.02 |
ENST00000295756.6
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr17_-_64225508 | 0.02 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr15_+_75491203 | 0.02 |
ENST00000562637.1
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr9_+_125132803 | 0.02 |
ENST00000540753.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr2_-_40680578 | 0.02 |
ENST00000455476.1
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr1_-_109618566 | 0.02 |
ENST00000338366.5
|
TAF13
|
TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa |
| chr19_-_19302931 | 0.02 |
ENST00000444486.3
ENST00000514819.3 ENST00000585679.1 ENST00000162023.5 |
MEF2BNB-MEF2B
MEF2BNB
MEF2B
|
MEF2BNB-MEF2B readthrough MEF2B neighbor myocyte enhancer factor 2B |
| chr1_-_190446759 | 0.02 |
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr3_+_158519654 | 0.02 |
ENST00000415822.2
ENST00000392813.4 ENST00000264266.8 |
MFSD1
|
major facilitator superfamily domain containing 1 |
| chr15_+_64680003 | 0.02 |
ENST00000261884.3
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr3_+_183853052 | 0.02 |
ENST00000273783.3
ENST00000432569.1 ENST00000444495.1 |
EIF2B5
|
eukaryotic translation initiation factor 2B, subunit 5 epsilon, 82kDa |
| chr3_-_101039402 | 0.02 |
ENST00000193391.7
|
IMPG2
|
interphotoreceptor matrix proteoglycan 2 |
| chr6_+_55192267 | 0.02 |
ENST00000340465.2
|
GFRAL
|
GDNF family receptor alpha like |
| chr3_+_50273625 | 0.02 |
ENST00000536647.1
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 |
| chr8_-_102803163 | 0.02 |
ENST00000523645.1
ENST00000520346.1 ENST00000220931.6 ENST00000522448.1 ENST00000522951.1 ENST00000522252.1 ENST00000519098.1 |
NCALD
|
neurocalcin delta |
| chr2_+_220363579 | 0.02 |
ENST00000313597.5
ENST00000373917.3 ENST00000358215.3 ENST00000373908.1 ENST00000455657.1 ENST00000435316.1 ENST00000341142.3 |
GMPPA
|
GDP-mannose pyrophosphorylase A |
| chr7_-_99716952 | 0.02 |
ENST00000523306.1
ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr6_-_127780510 | 0.02 |
ENST00000487331.2
ENST00000483725.3 |
KIAA0408
|
KIAA0408 |
| chr3_+_28390637 | 0.02 |
ENST00000420223.1
ENST00000383768.2 |
ZCWPW2
|
zinc finger, CW type with PWWP domain 2 |
| chr2_+_32288657 | 0.02 |
ENST00000345662.1
|
SPAST
|
spastin |
| chr21_-_35899113 | 0.02 |
ENST00000492600.1
ENST00000481448.1 ENST00000381132.2 |
RCAN1
|
regulator of calcineurin 1 |
| chr18_+_46065393 | 0.02 |
ENST00000256413.3
|
CTIF
|
CBP80/20-dependent translation initiation factor |
| chr19_-_19887185 | 0.02 |
ENST00000590092.1
ENST00000589910.1 ENST00000589508.1 ENST00000586645.1 ENST00000586816.1 ENST00000588223.1 ENST00000586885.1 |
LINC00663
|
long intergenic non-protein coding RNA 663 |
| chr12_-_71031220 | 0.02 |
ENST00000334414.6
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr12_+_20963632 | 0.02 |
ENST00000540853.1
ENST00000261196.2 |
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr4_+_95128748 | 0.02 |
ENST00000359052.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr5_+_55149150 | 0.02 |
ENST00000297015.3
|
IL31RA
|
interleukin 31 receptor A |
| chr3_+_108541608 | 0.02 |
ENST00000426646.1
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr15_+_90118723 | 0.02 |
ENST00000560985.1
|
TICRR
|
TOPBP1-interacting checkpoint and replication regulator |
| chr15_+_90118685 | 0.02 |
ENST00000268138.7
|
TICRR
|
TOPBP1-interacting checkpoint and replication regulator |
| chr2_-_208031943 | 0.02 |
ENST00000421199.1
ENST00000457962.1 |
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr6_-_110501200 | 0.02 |
ENST00000392586.1
ENST00000419252.1 ENST00000392589.1 ENST00000392588.1 ENST00000359451.2 |
WASF1
|
WAS protein family, member 1 |
| chr10_-_61900762 | 0.02 |
ENST00000355288.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr19_+_36632485 | 0.02 |
ENST00000586963.1
|
CAPNS1
|
calpain, small subunit 1 |
| chr10_-_5046042 | 0.02 |
ENST00000421196.3
ENST00000455190.1 |
AKR1C2
|
aldo-keto reductase family 1, member C2 |
| chr2_-_136678123 | 0.02 |
ENST00000422708.1
|
DARS
|
aspartyl-tRNA synthetase |
Network of associatons between targets according to the STRING database.
First level regulatory network of VAX1_GSX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.3 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.1 | GO:0051795 | positive regulation of catagen(GO:0051795) activation of meiosis(GO:0090427) |
| 0.0 | 0.1 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.0 | 0.1 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.1 | GO:0060489 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.0 | 0.1 | GO:0071877 | regulation of adrenergic receptor signaling pathway(GO:0071877) |
| 0.0 | 0.0 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.0 | 0.1 | GO:0010900 | negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 0.0 | 0.1 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.1 | GO:0090244 | negative regulation of B cell differentiation(GO:0045578) Wnt signaling pathway involved in somitogenesis(GO:0090244) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 0.0 | 0.1 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) |
| 0.0 | 0.6 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.1 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.0 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.1 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |