Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for UGGUCCC
Z-value: 0.62
miRNA associated with seed UGGUCCC
| Name | miRBASE accession |
|---|---|
|
hsa-miR-133a-3p.1
|
Activity profile of UGGUCCC motif
Sorted Z-values of UGGUCCC motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_41174424 | 0.40 |
ENST00000355653.3
|
VAT1
|
vesicle amine transport 1 |
| chr16_+_577697 | 0.34 |
ENST00000562370.1
ENST00000568988.1 ENST00000219611.2 |
CAPN15
|
calpain 15 |
| chr7_+_2671568 | 0.32 |
ENST00000258796.7
|
TTYH3
|
tweety family member 3 |
| chr7_+_100728720 | 0.28 |
ENST00000306085.6
ENST00000412507.1 |
TRIM56
|
tripartite motif containing 56 |
| chr2_+_28615669 | 0.27 |
ENST00000379619.1
ENST00000264716.4 |
FOSL2
|
FOS-like antigen 2 |
| chr19_+_15218180 | 0.27 |
ENST00000342784.2
ENST00000597977.1 ENST00000600440.1 |
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans) |
| chr11_+_66610883 | 0.25 |
ENST00000309657.3
ENST00000524506.1 |
RCE1
|
Ras converting CAAX endopeptidase 1 |
| chr19_-_10444188 | 0.24 |
ENST00000293677.6
|
RAVER1
|
ribonucleoprotein, PTB-binding 1 |
| chr19_-_663277 | 0.23 |
ENST00000292363.5
|
RNF126
|
ring finger protein 126 |
| chr19_+_49468558 | 0.21 |
ENST00000331825.6
|
FTL
|
ferritin, light polypeptide |
| chr16_+_58059470 | 0.20 |
ENST00000219271.3
|
MMP15
|
matrix metallopeptidase 15 (membrane-inserted) |
| chr17_-_1928621 | 0.20 |
ENST00000331238.6
|
RTN4RL1
|
reticulon 4 receptor-like 1 |
| chr16_+_29823552 | 0.20 |
ENST00000300797.6
|
PRRT2
|
proline-rich transmembrane protein 2 |
| chr19_-_4400415 | 0.20 |
ENST00000598564.1
ENST00000417295.2 ENST00000269886.3 |
SH3GL1
|
SH3-domain GRB2-like 1 |
| chr1_-_159893507 | 0.20 |
ENST00000368096.1
|
TAGLN2
|
transgelin 2 |
| chr19_+_17622415 | 0.19 |
ENST00000252603.2
ENST00000600923.1 |
PGLS
|
6-phosphogluconolactonase |
| chr14_+_23775971 | 0.18 |
ENST00000250405.5
|
BCL2L2
|
BCL2-like 2 |
| chr1_+_154975110 | 0.18 |
ENST00000535420.1
ENST00000368426.3 |
ZBTB7B
|
zinc finger and BTB domain containing 7B |
| chr22_+_21771656 | 0.18 |
ENST00000407464.2
|
HIC2
|
hypermethylated in cancer 2 |
| chr6_+_21593972 | 0.17 |
ENST00000244745.1
ENST00000543472.1 |
SOX4
|
SRY (sex determining region Y)-box 4 |
| chr2_+_85981008 | 0.17 |
ENST00000306279.3
|
ATOH8
|
atonal homolog 8 (Drosophila) |
| chr15_-_75135453 | 0.17 |
ENST00000569437.1
ENST00000440863.2 |
ULK3
|
unc-51 like kinase 3 |
| chr11_-_62414070 | 0.17 |
ENST00000540933.1
ENST00000346178.4 ENST00000356638.3 ENST00000534779.1 ENST00000525994.1 |
GANAB
|
glucosidase, alpha; neutral AB |
| chrX_-_153775426 | 0.16 |
ENST00000393562.2
|
G6PD
|
glucose-6-phosphate dehydrogenase |
| chr22_+_29469012 | 0.15 |
ENST00000400335.4
ENST00000400338.2 |
KREMEN1
|
kringle containing transmembrane protein 1 |
| chr3_-_138665969 | 0.15 |
ENST00000330315.3
|
FOXL2
|
forkhead box L2 |
| chr5_+_140855495 | 0.15 |
ENST00000308177.3
|
PCDHGC3
|
protocadherin gamma subfamily C, 3 |
| chr17_+_60704762 | 0.15 |
ENST00000303375.5
|
MRC2
|
mannose receptor, C type 2 |
| chr6_+_33168597 | 0.14 |
ENST00000374675.3
|
SLC39A7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr11_+_61520075 | 0.14 |
ENST00000278836.5
|
MYRF
|
myelin regulatory factor |
| chr17_+_37026106 | 0.14 |
ENST00000318008.6
|
LASP1
|
LIM and SH3 protein 1 |
| chr2_+_27593389 | 0.14 |
ENST00000233575.2
ENST00000543024.1 ENST00000537606.1 |
SNX17
|
sorting nexin 17 |
| chr9_-_21335356 | 0.14 |
ENST00000359039.4
|
KLHL9
|
kelch-like family member 9 |
| chr10_+_72238517 | 0.14 |
ENST00000263563.6
|
PALD1
|
phosphatase domain containing, paladin 1 |
| chr12_+_56511943 | 0.13 |
ENST00000257940.2
ENST00000552345.1 ENST00000551880.1 ENST00000546903.1 ENST00000551790.1 |
ZC3H10
ESYT1
|
zinc finger CCCH-type containing 10 extended synaptotagmin-like protein 1 |
| chr12_+_122516626 | 0.13 |
ENST00000319080.7
|
MLXIP
|
MLX interacting protein |
| chr7_+_28452130 | 0.13 |
ENST00000357727.2
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr5_+_140762268 | 0.13 |
ENST00000518325.1
|
PCDHGA7
|
protocadherin gamma subfamily A, 7 |
| chr1_+_36348790 | 0.13 |
ENST00000373204.4
|
AGO1
|
argonaute RISC catalytic component 1 |
| chr19_+_34287751 | 0.13 |
ENST00000590771.1
ENST00000589786.1 ENST00000284006.6 ENST00000588881.1 |
KCTD15
|
potassium channel tetramerization domain containing 15 |
| chr12_-_121342170 | 0.13 |
ENST00000353487.2
|
SPPL3
|
signal peptide peptidase like 3 |
| chr9_+_131549483 | 0.13 |
ENST00000372648.5
ENST00000539497.1 |
TBC1D13
|
TBC1 domain family, member 13 |
| chr20_+_36149602 | 0.13 |
ENST00000062104.2
ENST00000346199.2 |
NNAT
|
neuronatin |
| chr22_-_38669030 | 0.13 |
ENST00000361906.3
|
TMEM184B
|
transmembrane protein 184B |
| chr19_+_48216600 | 0.12 |
ENST00000263277.3
ENST00000538399.1 |
EHD2
|
EH-domain containing 2 |
| chr12_-_49504655 | 0.12 |
ENST00000551782.1
ENST00000267102.8 |
LMBR1L
|
limb development membrane protein 1-like |
| chr12_+_53574464 | 0.12 |
ENST00000416904.3
|
ZNF740
|
zinc finger protein 740 |
| chr16_-_66584059 | 0.12 |
ENST00000417693.3
ENST00000544898.1 ENST00000569718.1 ENST00000527284.1 ENST00000299697.7 ENST00000451102.2 |
TK2
|
thymidine kinase 2, mitochondrial |
| chr1_+_18958008 | 0.12 |
ENST00000420770.2
ENST00000400661.3 |
PAX7
|
paired box 7 |
| chr11_+_68816356 | 0.12 |
ENST00000294309.3
ENST00000542467.1 |
TPCN2
|
two pore segment channel 2 |
| chr2_-_166930131 | 0.12 |
ENST00000303395.4
ENST00000409050.1 ENST00000423058.2 ENST00000375405.3 |
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit |
| chr1_+_15736359 | 0.12 |
ENST00000375980.4
|
EFHD2
|
EF-hand domain family, member D2 |
| chr15_-_68521996 | 0.12 |
ENST00000418702.2
ENST00000565471.1 ENST00000564752.1 ENST00000566347.1 ENST00000249806.5 ENST00000562767.1 |
CLN6
RP11-315D16.2
|
ceroid-lipofuscinosis, neuronal 6, late infantile, variant Uncharacterized protein |
| chr19_+_1026298 | 0.11 |
ENST00000263097.4
|
CNN2
|
calponin 2 |
| chr22_-_39239987 | 0.11 |
ENST00000333039.2
|
NPTXR
|
neuronal pentraxin receptor |
| chr19_+_49660997 | 0.11 |
ENST00000598691.1
ENST00000252826.5 |
TRPM4
|
transient receptor potential cation channel, subfamily M, member 4 |
| chr9_+_130830451 | 0.10 |
ENST00000373068.2
ENST00000373069.5 |
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25 |
| chr17_+_72983674 | 0.10 |
ENST00000337231.5
|
CDR2L
|
cerebellar degeneration-related protein 2-like |
| chr12_-_54779511 | 0.10 |
ENST00000551109.1
ENST00000546970.1 |
ZNF385A
|
zinc finger protein 385A |
| chr11_-_75062730 | 0.10 |
ENST00000420843.2
ENST00000360025.3 |
ARRB1
|
arrestin, beta 1 |
| chr19_+_2164126 | 0.10 |
ENST00000398665.3
|
DOT1L
|
DOT1-like histone H3K79 methyltransferase |
| chr11_+_64794875 | 0.10 |
ENST00000377244.3
ENST00000534637.1 ENST00000524831.1 |
SNX15
|
sorting nexin 15 |
| chr12_-_50222187 | 0.10 |
ENST00000335999.6
|
NCKAP5L
|
NCK-associated protein 5-like |
| chr19_+_797392 | 0.10 |
ENST00000350092.4
ENST00000349038.4 ENST00000586481.1 ENST00000585535.1 |
PTBP1
|
polypyrimidine tract binding protein 1 |
| chr17_+_4736627 | 0.09 |
ENST00000355280.6
ENST00000347992.7 |
MINK1
|
misshapen-like kinase 1 |
| chr20_+_35201857 | 0.09 |
ENST00000373874.2
|
TGIF2
|
TGFB-induced factor homeobox 2 |
| chr20_-_30310336 | 0.09 |
ENST00000434194.1
ENST00000376062.2 |
BCL2L1
|
BCL2-like 1 |
| chr13_-_79177673 | 0.09 |
ENST00000377208.5
|
POU4F1
|
POU class 4 homeobox 1 |
| chr5_+_178286925 | 0.09 |
ENST00000322434.3
|
ZNF354B
|
zinc finger protein 354B |
| chr1_+_32645269 | 0.09 |
ENST00000373610.3
|
TXLNA
|
taxilin alpha |
| chr22_-_42017021 | 0.09 |
ENST00000263256.6
|
DESI1
|
desumoylating isopeptidase 1 |
| chr11_+_63706444 | 0.08 |
ENST00000377793.4
ENST00000456907.2 ENST00000539656.1 |
NAA40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chr17_-_27893990 | 0.08 |
ENST00000307201.4
|
ABHD15
|
abhydrolase domain containing 15 |
| chr22_+_20119320 | 0.08 |
ENST00000334554.7
ENST00000320602.7 ENST00000405930.3 |
ZDHHC8
|
zinc finger, DHHC-type containing 8 |
| chr17_-_40021656 | 0.08 |
ENST00000319121.3
|
KLHL11
|
kelch-like family member 11 |
| chr11_+_46639071 | 0.08 |
ENST00000580238.1
ENST00000581416.1 ENST00000529655.1 ENST00000533325.1 ENST00000581438.1 ENST00000583249.1 ENST00000530500.1 ENST00000526508.1 ENST00000578626.1 ENST00000577256.1 ENST00000524625.1 ENST00000582547.1 ENST00000359513.4 ENST00000528494.1 |
ATG13
|
autophagy related 13 |
| chr3_-_49823941 | 0.08 |
ENST00000321599.4
ENST00000395238.1 ENST00000468463.1 ENST00000460540.1 |
IP6K1
|
inositol hexakisphosphate kinase 1 |
| chr8_-_38326139 | 0.08 |
ENST00000335922.5
ENST00000532791.1 ENST00000397091.5 |
FGFR1
|
fibroblast growth factor receptor 1 |
| chr4_-_141075330 | 0.08 |
ENST00000509479.2
|
MAML3
|
mastermind-like 3 (Drosophila) |
| chr1_+_2985760 | 0.08 |
ENST00000378391.2
ENST00000514189.1 ENST00000270722.5 |
PRDM16
|
PR domain containing 16 |
| chr14_+_75348592 | 0.08 |
ENST00000334220.4
|
DLST
|
dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) |
| chr12_-_42538657 | 0.08 |
ENST00000398675.3
|
GXYLT1
|
glucoside xylosyltransferase 1 |
| chr1_+_16767167 | 0.08 |
ENST00000337132.5
|
NECAP2
|
NECAP endocytosis associated 2 |
| chr19_+_8429031 | 0.08 |
ENST00000301455.2
ENST00000541807.1 ENST00000393962.2 |
ANGPTL4
|
angiopoietin-like 4 |
| chr6_+_1610681 | 0.07 |
ENST00000380874.2
|
FOXC1
|
forkhead box C1 |
| chr19_-_7293942 | 0.07 |
ENST00000341500.5
ENST00000302850.5 |
INSR
|
insulin receptor |
| chr17_+_7210294 | 0.07 |
ENST00000336452.7
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chrX_-_100604184 | 0.07 |
ENST00000372902.3
|
TIMM8A
|
translocase of inner mitochondrial membrane 8 homolog A (yeast) |
| chr8_-_134309335 | 0.07 |
ENST00000522890.1
ENST00000323851.7 ENST00000518176.1 ENST00000354944.5 ENST00000537882.1 ENST00000522476.1 ENST00000518066.1 ENST00000521544.1 ENST00000518480.1 ENST00000523892.1 |
NDRG1
|
N-myc downstream regulated 1 |
| chr16_-_10674528 | 0.07 |
ENST00000359543.3
|
EMP2
|
epithelial membrane protein 2 |
| chr16_-_2390704 | 0.07 |
ENST00000301732.5
ENST00000382381.3 |
ABCA3
|
ATP-binding cassette, sub-family A (ABC1), member 3 |
| chr9_-_123639600 | 0.07 |
ENST00000373896.3
|
PHF19
|
PHD finger protein 19 |
| chr1_+_32573636 | 0.07 |
ENST00000373625.3
|
KPNA6
|
karyopherin alpha 6 (importin alpha 7) |
| chr17_-_48227877 | 0.07 |
ENST00000316878.6
|
PPP1R9B
|
protein phosphatase 1, regulatory subunit 9B |
| chr15_+_91411810 | 0.07 |
ENST00000268171.3
|
FURIN
|
furin (paired basic amino acid cleaving enzyme) |
| chr9_+_118916082 | 0.07 |
ENST00000328252.3
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1 |
| chr10_+_104474207 | 0.07 |
ENST00000602831.1
ENST00000369893.5 |
SFXN2
|
sideroflexin 2 |
| chr1_-_1709845 | 0.07 |
ENST00000341426.5
ENST00000344463.4 |
NADK
|
NAD kinase |
| chr10_-_75410771 | 0.07 |
ENST00000372873.4
|
SYNPO2L
|
synaptopodin 2-like |
| chr4_-_7873981 | 0.07 |
ENST00000360265.4
|
AFAP1
|
actin filament associated protein 1 |
| chr5_+_140810132 | 0.07 |
ENST00000252085.3
|
PCDHGA12
|
protocadherin gamma subfamily A, 12 |
| chr17_-_26733179 | 0.07 |
ENST00000440501.1
ENST00000321666.5 |
SLC46A1
|
solute carrier family 46 (folate transporter), member 1 |
| chrX_-_124097620 | 0.06 |
ENST00000371130.3
ENST00000422452.2 |
TENM1
|
teneurin transmembrane protein 1 |
| chr3_-_48723268 | 0.06 |
ENST00000439518.1
ENST00000416649.2 ENST00000341520.4 ENST00000294129.2 |
NCKIPSD
|
NCK interacting protein with SH3 domain |
| chr20_-_47444420 | 0.06 |
ENST00000371941.3
ENST00000396220.1 |
PREX1
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
| chr19_-_16682987 | 0.06 |
ENST00000431408.1
ENST00000436553.2 ENST00000595753.1 |
SLC35E1
|
solute carrier family 35, member E1 |
| chr20_+_56964169 | 0.06 |
ENST00000475243.1
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr2_-_26205340 | 0.06 |
ENST00000264712.3
|
KIF3C
|
kinesin family member 3C |
| chr6_+_41514078 | 0.06 |
ENST00000373063.3
ENST00000373060.1 |
FOXP4
|
forkhead box P4 |
| chr7_+_129710350 | 0.06 |
ENST00000335420.5
ENST00000463413.1 |
KLHDC10
|
kelch domain containing 10 |
| chr8_-_91658303 | 0.06 |
ENST00000458549.2
|
TMEM64
|
transmembrane protein 64 |
| chr19_+_38879061 | 0.06 |
ENST00000587013.1
|
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr5_+_140743859 | 0.06 |
ENST00000518069.1
|
PCDHGA5
|
protocadherin gamma subfamily A, 5 |
| chr12_-_133405288 | 0.05 |
ENST00000204726.3
|
GOLGA3
|
golgin A3 |
| chr12_-_57030115 | 0.05 |
ENST00000379441.3
ENST00000179765.5 ENST00000551812.1 |
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A |
| chr14_+_69658194 | 0.05 |
ENST00000409018.3
ENST00000409014.1 ENST00000409675.1 |
EXD2
|
exonuclease 3'-5' domain containing 2 |
| chrX_-_48901012 | 0.05 |
ENST00000315869.7
|
TFE3
|
transcription factor binding to IGHM enhancer 3 |
| chr6_-_134373732 | 0.05 |
ENST00000275230.5
|
SLC2A12
|
solute carrier family 2 (facilitated glucose transporter), member 12 |
| chr5_-_172198190 | 0.05 |
ENST00000239223.3
|
DUSP1
|
dual specificity phosphatase 1 |
| chr11_-_71791435 | 0.05 |
ENST00000351960.6
ENST00000541719.1 ENST00000535111.1 |
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr3_+_25469724 | 0.05 |
ENST00000437042.2
|
RARB
|
retinoic acid receptor, beta |
| chr17_-_37557846 | 0.05 |
ENST00000394294.3
ENST00000583610.1 ENST00000264658.6 |
FBXL20
|
F-box and leucine-rich repeat protein 20 |
| chr10_-_79686284 | 0.05 |
ENST00000372391.2
ENST00000372388.2 |
DLG5
|
discs, large homolog 5 (Drosophila) |
| chr2_-_208030647 | 0.05 |
ENST00000309446.6
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr5_-_175964366 | 0.05 |
ENST00000274811.4
|
RNF44
|
ring finger protein 44 |
| chr20_-_32308028 | 0.05 |
ENST00000409299.3
ENST00000217398.3 ENST00000344022.3 |
PXMP4
|
peroxisomal membrane protein 4, 24kDa |
| chr14_-_74253948 | 0.05 |
ENST00000394071.2
|
ELMSAN1
|
ELM2 and Myb/SANT-like domain containing 1 |
| chr22_-_44894178 | 0.05 |
ENST00000341255.3
|
LDOC1L
|
leucine zipper, down-regulated in cancer 1-like |
| chr2_+_33661382 | 0.05 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr5_+_140739537 | 0.05 |
ENST00000522605.1
|
PCDHGB2
|
protocadherin gamma subfamily B, 2 |
| chr7_+_1727755 | 0.05 |
ENST00000424383.2
|
ELFN1
|
extracellular leucine-rich repeat and fibronectin type III domain containing 1 |
| chr3_+_14444063 | 0.05 |
ENST00000454876.2
ENST00000360861.3 ENST00000416216.2 |
SLC6A6
|
solute carrier family 6 (neurotransmitter transporter), member 6 |
| chr5_+_140772381 | 0.05 |
ENST00000398604.2
|
PCDHGA8
|
protocadherin gamma subfamily A, 8 |
| chr6_+_42952237 | 0.04 |
ENST00000485511.1
ENST00000394110.3 ENST00000472118.1 ENST00000461010.1 |
PPP2R5D
|
protein phosphatase 2, regulatory subunit B', delta |
| chr14_-_75179774 | 0.04 |
ENST00000555249.1
ENST00000556202.1 ENST00000356357.4 ENST00000338772.5 |
AREL1
AC007956.1
|
apoptosis resistant E3 ubiquitin protein ligase 1 Full-length cDNA 5-PRIME end of clone CS0CAP004YO05 of Thymus of Homo sapiens (human); Uncharacterized protein |
| chr12_+_58148842 | 0.04 |
ENST00000266643.5
|
MARCH9
|
membrane-associated ring finger (C3HC4) 9 |
| chr5_+_140792614 | 0.04 |
ENST00000398610.2
|
PCDHGA10
|
protocadherin gamma subfamily A, 10 |
| chr5_+_140767452 | 0.04 |
ENST00000519479.1
|
PCDHGB4
|
protocadherin gamma subfamily B, 4 |
| chr17_-_8066250 | 0.04 |
ENST00000488857.1
ENST00000481878.1 ENST00000316509.6 ENST00000498285.1 |
VAMP2
RP11-599B13.6
|
vesicle-associated membrane protein 2 (synaptobrevin 2) Uncharacterized protein |
| chr5_+_140753444 | 0.04 |
ENST00000517434.1
|
PCDHGA6
|
protocadherin gamma subfamily A, 6 |
| chr9_+_131843377 | 0.04 |
ENST00000372546.4
ENST00000406974.3 ENST00000540102.1 |
DOLPP1
|
dolichyldiphosphatase 1 |
| chr2_-_73298802 | 0.04 |
ENST00000411783.1
ENST00000410065.1 ENST00000442582.1 ENST00000272433.2 |
SFXN5
|
sideroflexin 5 |
| chr1_-_150947343 | 0.04 |
ENST00000271688.6
ENST00000368954.5 |
CERS2
|
ceramide synthase 2 |
| chr1_+_151043070 | 0.04 |
ENST00000368918.3
ENST00000368917.1 |
GABPB2
|
GA binding protein transcription factor, beta subunit 2 |
| chr5_+_140800638 | 0.04 |
ENST00000398587.2
ENST00000518882.1 |
PCDHGA11
|
protocadherin gamma subfamily A, 11 |
| chr19_-_14629224 | 0.04 |
ENST00000254322.2
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr15_-_37390482 | 0.04 |
ENST00000559085.1
ENST00000397624.3 |
MEIS2
|
Meis homeobox 2 |
| chr11_+_818902 | 0.03 |
ENST00000336615.4
|
PNPLA2
|
patatin-like phospholipase domain containing 2 |
| chr18_+_905104 | 0.03 |
ENST00000579794.1
|
ADCYAP1
|
adenylate cyclase activating polypeptide 1 (pituitary) |
| chr5_+_140749803 | 0.03 |
ENST00000576222.1
|
PCDHGB3
|
protocadherin gamma subfamily B, 3 |
| chr1_+_165796753 | 0.03 |
ENST00000367879.4
|
UCK2
|
uridine-cytidine kinase 2 |
| chr9_+_109625378 | 0.03 |
ENST00000277225.5
ENST00000457913.1 ENST00000472574.1 |
ZNF462
|
zinc finger protein 462 |
| chr20_-_44718538 | 0.03 |
ENST00000290231.6
ENST00000372291.3 |
NCOA5
|
nuclear receptor coactivator 5 |
| chr5_+_140718396 | 0.03 |
ENST00000394576.2
|
PCDHGA2
|
protocadherin gamma subfamily A, 2 |
| chr5_+_140729649 | 0.03 |
ENST00000523390.1
|
PCDHGB1
|
protocadherin gamma subfamily B, 1 |
| chr5_+_140797296 | 0.03 |
ENST00000398594.2
|
PCDHGB7
|
protocadherin gamma subfamily B, 7 |
| chr17_-_74733404 | 0.03 |
ENST00000508921.3
ENST00000583836.1 ENST00000358156.6 ENST00000392485.2 ENST00000359995.5 |
SRSF2
|
serine/arginine-rich splicing factor 2 |
| chr7_+_140774032 | 0.03 |
ENST00000565468.1
|
TMEM178B
|
transmembrane protein 178B |
| chr5_+_140787600 | 0.03 |
ENST00000520790.1
|
PCDHGB6
|
protocadherin gamma subfamily B, 6 |
| chr16_+_50775948 | 0.03 |
ENST00000569681.1
ENST00000569418.1 ENST00000540145.1 |
CYLD
|
cylindromatosis (turban tumor syndrome) |
| chr2_-_55844720 | 0.03 |
ENST00000345102.5
ENST00000272313.5 ENST00000407823.3 |
SMEK2
|
SMEK homolog 2, suppressor of mek1 (Dictyostelium) |
| chr10_+_101419187 | 0.03 |
ENST00000370489.4
|
ENTPD7
|
ectonucleoside triphosphate diphosphohydrolase 7 |
| chr17_+_7788104 | 0.03 |
ENST00000380358.4
|
CHD3
|
chromodomain helicase DNA binding protein 3 |
| chr6_+_107811162 | 0.03 |
ENST00000317357.5
|
SOBP
|
sine oculis binding protein homolog (Drosophila) |
| chr5_-_133561752 | 0.03 |
ENST00000519718.1
ENST00000481195.1 |
CTD-2410N18.5
PPP2CA
|
S-phase kinase-associated protein 1 protein phosphatase 2, catalytic subunit, alpha isozyme |
| chr4_-_89744457 | 0.02 |
ENST00000395002.2
|
FAM13A
|
family with sequence similarity 13, member A |
| chr14_+_74111578 | 0.02 |
ENST00000554113.1
ENST00000555631.2 ENST00000553645.2 ENST00000311089.3 ENST00000555919.3 ENST00000554339.1 ENST00000554871.1 |
DNAL1
|
dynein, axonemal, light chain 1 |
| chr5_-_37839782 | 0.02 |
ENST00000326524.2
ENST00000515058.1 |
GDNF
|
glial cell derived neurotrophic factor |
| chr8_-_103876965 | 0.02 |
ENST00000337198.5
|
AZIN1
|
antizyme inhibitor 1 |
| chr5_+_179159813 | 0.02 |
ENST00000292599.3
|
MAML1
|
mastermind-like 1 (Drosophila) |
| chr1_+_45265897 | 0.02 |
ENST00000372201.4
|
PLK3
|
polo-like kinase 3 |
| chr12_-_117175819 | 0.02 |
ENST00000261318.3
ENST00000536380.1 |
C12orf49
|
chromosome 12 open reading frame 49 |
| chr12_+_10365404 | 0.02 |
ENST00000266458.5
ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr6_-_52441713 | 0.02 |
ENST00000182527.3
|
TRAM2
|
translocation associated membrane protein 2 |
| chr1_-_179198702 | 0.02 |
ENST00000502732.1
|
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr19_+_10828724 | 0.02 |
ENST00000585892.1
ENST00000314646.5 ENST00000359692.6 |
DNM2
|
dynamin 2 |
| chr11_+_64009072 | 0.02 |
ENST00000535135.1
ENST00000394540.3 |
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr7_+_114055052 | 0.02 |
ENST00000462331.1
ENST00000408937.3 ENST00000403559.4 ENST00000350908.4 ENST00000393498.2 ENST00000393495.3 ENST00000378237.3 ENST00000393489.3 |
FOXP2
|
forkhead box P2 |
| chr11_-_6502534 | 0.02 |
ENST00000254584.2
ENST00000525235.1 ENST00000445086.2 |
ARFIP2
|
ADP-ribosylation factor interacting protein 2 |
| chr5_+_140710061 | 0.02 |
ENST00000517417.1
ENST00000378105.3 |
PCDHGA1
|
protocadherin gamma subfamily A, 1 |
| chr1_-_155942086 | 0.02 |
ENST00000368315.4
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
| chr1_+_162039558 | 0.02 |
ENST00000530878.1
ENST00000361897.5 |
NOS1AP
|
nitric oxide synthase 1 (neuronal) adaptor protein |
| chr11_+_57520715 | 0.02 |
ENST00000524630.1
ENST00000529919.1 ENST00000399039.4 ENST00000533189.1 |
CTNND1
|
catenin (cadherin-associated protein), delta 1 |
| chr17_-_1359443 | 0.02 |
ENST00000574295.1
ENST00000398970.5 ENST00000300574.2 |
CRK
|
v-crk avian sarcoma virus CT10 oncogene homolog |
| chr15_+_90744533 | 0.02 |
ENST00000411539.2
|
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr12_+_50898881 | 0.02 |
ENST00000301180.5
|
DIP2B
|
DIP2 disco-interacting protein 2 homolog B (Drosophila) |
| chr17_-_27224621 | 0.02 |
ENST00000394906.2
ENST00000585169.1 ENST00000394908.4 |
FLOT2
|
flotillin 2 |
| chr1_-_149900122 | 0.02 |
ENST00000271628.8
|
SF3B4
|
splicing factor 3b, subunit 4, 49kDa |
| chr22_-_29784519 | 0.02 |
ENST00000357586.2
ENST00000356015.2 ENST00000432560.2 ENST00000317368.7 |
AP1B1
|
adaptor-related protein complex 1, beta 1 subunit |
| chr5_+_140723601 | 0.02 |
ENST00000253812.6
|
PCDHGA3
|
protocadherin gamma subfamily A, 3 |
| chr3_+_185303962 | 0.02 |
ENST00000296257.5
|
SENP2
|
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr8_+_143530791 | 0.02 |
ENST00000517894.1
|
BAI1
|
brain-specific angiogenesis inhibitor 1 |
| chr10_+_6186847 | 0.02 |
ENST00000536985.1
ENST00000379789.4 |
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr3_+_126423045 | 0.02 |
ENST00000290913.3
ENST00000508789.1 |
CHCHD6
|
coiled-coil-helix-coiled-coil-helix domain containing 6 |
| chr22_-_38902325 | 0.01 |
ENST00000396821.3
ENST00000381633.3 |
DDX17
|
DEAD (Asp-Glu-Ala-Asp) box helicase 17 |
| chr7_+_39989611 | 0.01 |
ENST00000181839.4
|
CDK13
|
cyclin-dependent kinase 13 |
| chr5_-_16509101 | 0.01 |
ENST00000399793.2
|
FAM134B
|
family with sequence similarity 134, member B |
| chr10_-_120514720 | 0.01 |
ENST00000369151.3
ENST00000340214.4 |
CACUL1
|
CDK2-associated, cullin domain 1 |
| chr5_+_141303373 | 0.01 |
ENST00000432126.2
ENST00000194118.4 |
KIAA0141
|
KIAA0141 |
| chr2_-_40679186 | 0.01 |
ENST00000406785.2
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
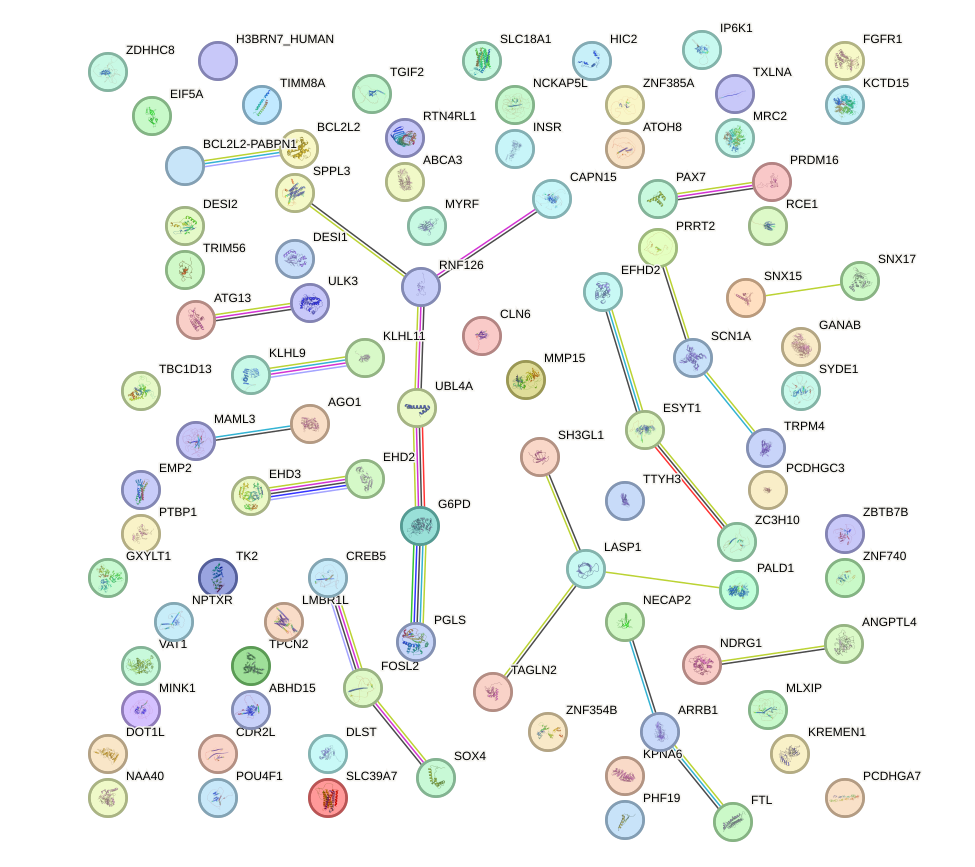
Network of associatons between targets according to the STRING database.
First level regulatory network of UGGUCCC
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.1 | 0.2 | GO:0071629 | cytoplasm-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071629) |
| 0.1 | 0.3 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.1 | 0.4 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.1 | 0.4 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.2 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.2 | GO:0003357 | noradrenergic neuron differentiation(GO:0003357) |
| 0.0 | 0.1 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.0 | 0.2 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.2 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.1 | GO:1904199 | positive regulation of regulation of vascular smooth muscle cell membrane depolarization(GO:1904199) regulation of vascular smooth muscle cell membrane depolarization(GO:1990736) |
| 0.0 | 0.1 | GO:2000830 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.0 | 0.1 | GO:1902256 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.0 | 0.1 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.0 | 0.1 | GO:0019082 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.0 | 0.1 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.0 | 0.1 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.1 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.0 | 0.3 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.0 | 0.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.1 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.1 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.1 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.1 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.1 | GO:1902162 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.1 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.1 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.0 | 0.2 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.0 | 0.1 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.1 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.1 | GO:0051958 | methotrexate transport(GO:0051958) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.1 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0008043 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0097232 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.1 | 0.2 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.1 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.0 | 0.1 | GO:0052836 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.0 | 0.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.2 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0015350 | methotrexate transporter activity(GO:0015350) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |