Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for UGAAAUG
Z-value: 0.55
Activity profile of UGAAAUG motif
Sorted Z-values of UGAAAUG motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_128003606 | 0.10 |
ENST00000324460.6
|
HSPA5
|
heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) |
| chr20_+_56884752 | 0.09 |
ENST00000244040.3
|
RAB22A
|
RAB22A, member RAS oncogene family |
| chr8_+_95732095 | 0.09 |
ENST00000414645.2
|
DPY19L4
|
dpy-19-like 4 (C. elegans) |
| chr11_+_95523621 | 0.09 |
ENST00000325542.5
ENST00000325486.5 ENST00000544522.1 ENST00000541365.1 |
CEP57
|
centrosomal protein 57kDa |
| chr5_-_65017921 | 0.09 |
ENST00000381007.4
|
SGTB
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr10_+_76586348 | 0.09 |
ENST00000372724.1
ENST00000287239.4 ENST00000372714.1 |
KAT6B
|
K(lysine) acetyltransferase 6B |
| chr4_-_80994210 | 0.09 |
ENST00000403729.2
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr1_+_112162381 | 0.08 |
ENST00000433097.1
ENST00000369709.3 ENST00000436150.2 |
RAP1A
|
RAP1A, member of RAS oncogene family |
| chr8_-_25315905 | 0.08 |
ENST00000221200.4
|
KCTD9
|
potassium channel tetramerization domain containing 9 |
| chr14_-_50319758 | 0.08 |
ENST00000298310.5
|
NEMF
|
nuclear export mediator factor |
| chr15_+_27111510 | 0.08 |
ENST00000335625.5
|
GABRA5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr5_+_52285144 | 0.08 |
ENST00000296585.5
|
ITGA2
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) |
| chr5_-_89770582 | 0.08 |
ENST00000316610.6
|
MBLAC2
|
metallo-beta-lactamase domain containing 2 |
| chr2_+_14772810 | 0.08 |
ENST00000295092.2
ENST00000331243.4 |
FAM84A
|
family with sequence similarity 84, member A |
| chr18_+_52495426 | 0.07 |
ENST00000262094.5
|
RAB27B
|
RAB27B, member RAS oncogene family |
| chr11_-_89956461 | 0.07 |
ENST00000320585.6
|
CHORDC1
|
cysteine and histidine-rich domain (CHORD) containing 1 |
| chrX_-_77395186 | 0.07 |
ENST00000341864.5
|
TAF9B
|
TAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31kDa |
| chr2_-_179343268 | 0.07 |
ENST00000424785.2
|
FKBP7
|
FK506 binding protein 7 |
| chr8_+_94712732 | 0.07 |
ENST00000518322.1
|
FAM92A1
|
family with sequence similarity 92, member A1 |
| chr1_-_85462623 | 0.07 |
ENST00000370608.3
|
MCOLN2
|
mucolipin 2 |
| chr11_+_107461948 | 0.06 |
ENST00000265840.7
ENST00000443271.2 |
ELMOD1
|
ELMO/CED-12 domain containing 1 |
| chr12_-_76425368 | 0.06 |
ENST00000602540.1
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1 |
| chr19_+_30433110 | 0.06 |
ENST00000542441.2
ENST00000392271.1 |
URI1
|
URI1, prefoldin-like chaperone |
| chr20_-_49575081 | 0.06 |
ENST00000371588.5
ENST00000371582.4 |
DPM1
|
dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit |
| chr1_+_6845497 | 0.06 |
ENST00000473578.1
ENST00000557126.1 |
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr9_+_114659046 | 0.06 |
ENST00000374279.3
|
UGCG
|
UDP-glucose ceramide glucosyltransferase |
| chrX_-_102942961 | 0.06 |
ENST00000434230.1
ENST00000418819.1 ENST00000360458.1 |
MORF4L2
|
mortality factor 4 like 2 |
| chr1_+_35734562 | 0.06 |
ENST00000314607.6
ENST00000373297.2 |
ZMYM4
|
zinc finger, MYM-type 4 |
| chrX_-_41782249 | 0.06 |
ENST00000442742.2
ENST00000421587.2 ENST00000378166.4 ENST00000318588.9 ENST00000361962.4 |
CASK
|
calcium/calmodulin-dependent serine protein kinase (MAGUK family) |
| chr1_-_70671216 | 0.06 |
ENST00000370952.3
|
LRRC40
|
leucine rich repeat containing 40 |
| chr4_+_123747834 | 0.06 |
ENST00000264498.3
|
FGF2
|
fibroblast growth factor 2 (basic) |
| chr9_-_94186131 | 0.06 |
ENST00000297689.3
|
NFIL3
|
nuclear factor, interleukin 3 regulated |
| chr3_-_171178157 | 0.06 |
ENST00000465393.1
ENST00000436636.2 ENST00000369326.5 ENST00000538048.1 ENST00000341852.6 |
TNIK
|
TRAF2 and NCK interacting kinase |
| chr10_+_111967345 | 0.06 |
ENST00000332674.5
ENST00000453116.1 |
MXI1
|
MAX interactor 1, dimerization protein |
| chr17_+_45727204 | 0.05 |
ENST00000290158.4
|
KPNB1
|
karyopherin (importin) beta 1 |
| chr1_-_23670817 | 0.05 |
ENST00000478691.1
|
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R |
| chr2_+_42795651 | 0.05 |
ENST00000407270.3
|
MTA3
|
metastasis associated 1 family, member 3 |
| chr14_+_102196739 | 0.05 |
ENST00000556973.1
|
RP11-796G6.2
|
Uncharacterized protein |
| chr12_-_109125285 | 0.05 |
ENST00000552871.1
ENST00000261401.3 |
CORO1C
|
coronin, actin binding protein, 1C |
| chr12_+_120875910 | 0.05 |
ENST00000551806.1
|
AL021546.6
|
Glutamyl-tRNA(Gln) amidotransferase subunit C, mitochondrial |
| chr2_-_180129484 | 0.05 |
ENST00000428443.3
|
SESTD1
|
SEC14 and spectrin domains 1 |
| chr6_+_71998506 | 0.05 |
ENST00000370435.4
|
OGFRL1
|
opioid growth factor receptor-like 1 |
| chr12_-_102513843 | 0.05 |
ENST00000551744.2
ENST00000552283.1 |
NUP37
|
nucleoporin 37kDa |
| chr7_+_21467642 | 0.05 |
ENST00000222584.3
ENST00000432066.2 |
SP4
|
Sp4 transcription factor |
| chr5_-_9546180 | 0.05 |
ENST00000382496.5
|
SEMA5A
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr15_-_79383102 | 0.05 |
ENST00000558480.2
ENST00000419573.3 |
RASGRF1
|
Ras protein-specific guanine nucleotide-releasing factor 1 |
| chr8_+_94767072 | 0.05 |
ENST00000452276.1
ENST00000453321.3 ENST00000498673.1 ENST00000518319.1 |
TMEM67
|
transmembrane protein 67 |
| chr14_+_73525144 | 0.05 |
ENST00000261973.7
ENST00000540173.1 |
RBM25
|
RNA binding motif protein 25 |
| chr20_+_39657454 | 0.05 |
ENST00000361337.2
|
TOP1
|
topoisomerase (DNA) I |
| chr7_-_27239703 | 0.05 |
ENST00000222753.4
|
HOXA13
|
homeobox A13 |
| chr5_-_59189545 | 0.05 |
ENST00000340635.6
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr3_+_178866199 | 0.05 |
ENST00000263967.3
|
PIK3CA
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
| chr14_-_39639523 | 0.05 |
ENST00000330149.5
ENST00000554018.1 ENST00000347691.5 |
TRAPPC6B
|
trafficking protein particle complex 6B |
| chr3_-_170303845 | 0.05 |
ENST00000231706.5
|
SLC7A14
|
solute carrier family 7, member 14 |
| chr2_+_69969106 | 0.05 |
ENST00000409920.1
ENST00000394295.4 ENST00000536030.1 |
ANXA4
|
annexin A4 |
| chrX_+_16804544 | 0.05 |
ENST00000380122.5
ENST00000398155.4 |
TXLNG
|
taxilin gamma |
| chr17_-_16118835 | 0.05 |
ENST00000582357.1
ENST00000436828.1 ENST00000411510.1 ENST00000268712.3 |
NCOR1
|
nuclear receptor corepressor 1 |
| chr12_+_27485823 | 0.05 |
ENST00000395901.2
ENST00000546179.1 |
ARNTL2
|
aryl hydrocarbon receptor nuclear translocator-like 2 |
| chr8_+_16884740 | 0.05 |
ENST00000318063.5
|
MICU3
|
mitochondrial calcium uptake family, member 3 |
| chr2_+_231577532 | 0.04 |
ENST00000258418.5
|
CAB39
|
calcium binding protein 39 |
| chr5_-_39425068 | 0.04 |
ENST00000515700.1
ENST00000339788.6 |
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr3_-_123304017 | 0.04 |
ENST00000383657.5
|
PTPLB
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member b |
| chrX_-_48980098 | 0.04 |
ENST00000156109.5
|
GPKOW
|
G patch domain and KOW motifs |
| chr13_+_95254085 | 0.04 |
ENST00000376958.4
|
GPR180
|
G protein-coupled receptor 180 |
| chr4_-_99851766 | 0.04 |
ENST00000450253.2
|
EIF4E
|
eukaryotic translation initiation factor 4E |
| chr10_+_105036909 | 0.04 |
ENST00000369849.4
|
INA
|
internexin neuronal intermediate filament protein, alpha |
| chr6_+_143772060 | 0.04 |
ENST00000367591.4
|
PEX3
|
peroxisomal biogenesis factor 3 |
| chr6_+_64281906 | 0.04 |
ENST00000370651.3
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr5_+_56205081 | 0.04 |
ENST00000285947.2
ENST00000541720.1 |
SETD9
|
SET domain containing 9 |
| chr22_+_42394780 | 0.04 |
ENST00000328823.9
|
WBP2NL
|
WBP2 N-terminal like |
| chr11_+_34073195 | 0.04 |
ENST00000341394.4
|
CAPRIN1
|
cell cycle associated protein 1 |
| chr7_-_83824169 | 0.04 |
ENST00000265362.4
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr15_+_79165151 | 0.04 |
ENST00000331268.5
|
MORF4L1
|
mortality factor 4 like 1 |
| chr8_-_41166953 | 0.04 |
ENST00000220772.3
|
SFRP1
|
secreted frizzled-related protein 1 |
| chr2_+_223725652 | 0.04 |
ENST00000357430.3
ENST00000392066.3 |
ACSL3
|
acyl-CoA synthetase long-chain family member 3 |
| chr14_+_96829814 | 0.04 |
ENST00000555181.1
ENST00000553699.1 ENST00000554182.1 |
GSKIP
|
GSK3B interacting protein |
| chr2_+_112895939 | 0.04 |
ENST00000331203.2
ENST00000409903.1 ENST00000409667.3 ENST00000409450.3 |
FBLN7
|
fibulin 7 |
| chr4_+_52709229 | 0.04 |
ENST00000334635.5
ENST00000381441.3 ENST00000381437.4 |
DCUN1D4
|
DCN1, defective in cullin neddylation 1, domain containing 4 |
| chr17_+_80477571 | 0.04 |
ENST00000335255.5
|
FOXK2
|
forkhead box K2 |
| chr5_+_122181184 | 0.04 |
ENST00000513881.1
|
SNX24
|
sorting nexin 24 |
| chr10_-_105677886 | 0.04 |
ENST00000224950.3
|
OBFC1
|
oligonucleotide/oligosaccharide-binding fold containing 1 |
| chr5_-_95297678 | 0.04 |
ENST00000237853.4
|
ELL2
|
elongation factor, RNA polymerase II, 2 |
| chr2_-_9770706 | 0.04 |
ENST00000381844.4
|
YWHAQ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta |
| chr11_+_10471836 | 0.04 |
ENST00000444303.2
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr7_-_107204337 | 0.04 |
ENST00000605888.1
ENST00000347053.3 |
COG5
|
component of oligomeric golgi complex 5 |
| chr7_-_108166505 | 0.04 |
ENST00000426128.2
ENST00000427008.1 ENST00000388728.5 ENST00000257694.8 ENST00000422087.1 ENST00000453144.1 ENST00000436062.1 |
PNPLA8
|
patatin-like phospholipase domain containing 8 |
| chr18_-_21977748 | 0.04 |
ENST00000399441.4
ENST00000319481.3 |
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr5_-_132299313 | 0.04 |
ENST00000265343.5
|
AFF4
|
AF4/FMR2 family, member 4 |
| chr8_-_81083731 | 0.04 |
ENST00000379096.5
|
TPD52
|
tumor protein D52 |
| chr13_+_97874574 | 0.04 |
ENST00000343600.4
ENST00000345429.6 ENST00000376673.3 |
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr11_+_120894781 | 0.04 |
ENST00000529397.1
ENST00000528512.1 ENST00000422003.2 |
TBCEL
|
tubulin folding cofactor E-like |
| chr6_+_167412665 | 0.04 |
ENST00000366847.4
|
FGFR1OP
|
FGFR1 oncogene partner |
| chr3_+_182971018 | 0.04 |
ENST00000326505.3
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr5_+_145583156 | 0.04 |
ENST00000265271.5
|
RBM27
|
RNA binding motif protein 27 |
| chr14_-_58894332 | 0.04 |
ENST00000395159.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr18_+_9913977 | 0.04 |
ENST00000400000.2
ENST00000340541.4 |
VAPA
|
VAMP (vesicle-associated membrane protein)-associated protein A, 33kDa |
| chr12_+_118814344 | 0.03 |
ENST00000397564.2
|
SUDS3
|
suppressor of defective silencing 3 homolog (S. cerevisiae) |
| chr13_-_21476900 | 0.03 |
ENST00000400602.2
ENST00000255305.6 |
XPO4
|
exportin 4 |
| chr14_-_74485960 | 0.03 |
ENST00000556242.1
ENST00000334696.6 |
ENTPD5
|
ectonucleoside triphosphate diphosphohydrolase 5 |
| chr11_+_13299186 | 0.03 |
ENST00000527998.1
ENST00000396441.3 ENST00000533520.1 ENST00000529825.1 ENST00000389707.4 ENST00000401424.1 ENST00000529388.1 ENST00000530357.1 ENST00000403290.1 ENST00000361003.4 ENST00000389708.3 ENST00000403510.3 ENST00000482049.1 |
ARNTL
|
aryl hydrocarbon receptor nuclear translocator-like |
| chr17_-_61905005 | 0.03 |
ENST00000584574.1
ENST00000585145.1 ENST00000427159.2 |
FTSJ3
|
FtsJ homolog 3 (E. coli) |
| chr13_-_45915221 | 0.03 |
ENST00000309246.5
ENST00000379060.4 ENST00000379055.1 ENST00000527226.1 ENST00000379056.1 |
TPT1
|
tumor protein, translationally-controlled 1 |
| chr3_+_196295482 | 0.03 |
ENST00000440469.1
ENST00000311630.6 |
FBXO45
|
F-box protein 45 |
| chr4_+_155665123 | 0.03 |
ENST00000336356.3
|
LRAT
|
lecithin retinol acyltransferase (phosphatidylcholine--retinol O-acyltransferase) |
| chr6_+_42749759 | 0.03 |
ENST00000314073.5
|
GLTSCR1L
|
GLTSCR1-like |
| chr2_-_169887827 | 0.03 |
ENST00000263817.6
|
ABCB11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
| chr3_+_191046810 | 0.03 |
ENST00000392455.3
ENST00000392456.3 |
CCDC50
|
coiled-coil domain containing 50 |
| chr13_-_22033392 | 0.03 |
ENST00000320220.9
ENST00000415724.1 ENST00000422251.1 ENST00000382466.3 ENST00000542645.1 ENST00000400590.3 |
ZDHHC20
|
zinc finger, DHHC-type containing 20 |
| chr2_-_136743169 | 0.03 |
ENST00000264161.4
|
DARS
|
aspartyl-tRNA synthetase |
| chr7_+_17338239 | 0.03 |
ENST00000242057.4
|
AHR
|
aryl hydrocarbon receptor |
| chr5_-_179719048 | 0.03 |
ENST00000523583.1
ENST00000393360.3 ENST00000455781.1 ENST00000347470.4 ENST00000452135.2 ENST00000343111.6 |
MAPK9
|
mitogen-activated protein kinase 9 |
| chr12_-_110318263 | 0.03 |
ENST00000318348.4
|
GLTP
|
glycolipid transfer protein |
| chr8_-_41909496 | 0.03 |
ENST00000265713.2
ENST00000406337.1 ENST00000396930.3 ENST00000485568.1 ENST00000426524.1 |
KAT6A
|
K(lysine) acetyltransferase 6A |
| chr1_+_210406121 | 0.03 |
ENST00000367012.3
|
SERTAD4
|
SERTA domain containing 4 |
| chr14_-_53019211 | 0.03 |
ENST00000557374.1
ENST00000281741.4 |
TXNDC16
|
thioredoxin domain containing 16 |
| chr1_+_213123915 | 0.03 |
ENST00000366968.4
ENST00000490792.1 |
VASH2
|
vasohibin 2 |
| chr6_-_52859046 | 0.03 |
ENST00000457564.1
ENST00000541324.1 ENST00000370960.1 |
GSTA4
|
glutathione S-transferase alpha 4 |
| chr10_+_98592009 | 0.03 |
ENST00000540664.1
ENST00000371103.3 |
LCOR
|
ligand dependent nuclear receptor corepressor |
| chr12_+_64845660 | 0.03 |
ENST00000331710.5
|
TBK1
|
TANK-binding kinase 1 |
| chr1_-_203320617 | 0.03 |
ENST00000354955.4
|
FMOD
|
fibromodulin |
| chrX_-_83757399 | 0.03 |
ENST00000373177.2
ENST00000297977.5 ENST00000506585.2 ENST00000449553.2 |
HDX
|
highly divergent homeobox |
| chr11_+_9406169 | 0.03 |
ENST00000379719.3
ENST00000527431.1 |
IPO7
|
importin 7 |
| chr1_+_65210772 | 0.03 |
ENST00000371072.4
ENST00000294428.3 |
RAVER2
|
ribonucleoprotein, PTB-binding 2 |
| chr2_+_148778570 | 0.03 |
ENST00000407073.1
|
MBD5
|
methyl-CpG binding domain protein 5 |
| chr3_+_152017181 | 0.03 |
ENST00000498502.1
ENST00000324196.5 ENST00000545754.1 ENST00000357472.3 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr2_+_26467762 | 0.03 |
ENST00000317799.5
ENST00000405867.3 ENST00000537713.1 |
HADHB
|
hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), beta subunit |
| chrX_-_15872914 | 0.03 |
ENST00000380291.1
ENST00000545766.1 ENST00000421527.2 ENST00000329235.2 |
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr1_+_101361626 | 0.03 |
ENST00000370112.4
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr15_-_27018175 | 0.03 |
ENST00000311550.5
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr11_+_108093839 | 0.03 |
ENST00000452508.2
|
ATM
|
ataxia telangiectasia mutated |
| chr17_+_47653178 | 0.03 |
ENST00000328741.5
|
NXPH3
|
neurexophilin 3 |
| chr16_-_53737795 | 0.03 |
ENST00000262135.4
ENST00000564374.1 ENST00000566096.1 |
RPGRIP1L
|
RPGRIP1-like |
| chr11_-_105892937 | 0.03 |
ENST00000301919.4
ENST00000534458.1 ENST00000530108.1 ENST00000530788.1 |
MSANTD4
|
Myb/SANT-like DNA-binding domain containing 4 with coiled-coils |
| chr1_+_181452678 | 0.03 |
ENST00000367570.1
ENST00000526775.1 ENST00000357570.5 ENST00000358338.5 ENST00000367567.4 |
CACNA1E
|
calcium channel, voltage-dependent, R type, alpha 1E subunit |
| chr5_+_79783788 | 0.03 |
ENST00000282226.4
|
FAM151B
|
family with sequence similarity 151, member B |
| chr2_-_54087066 | 0.03 |
ENST00000394705.2
ENST00000352846.3 ENST00000406625.2 |
GPR75
GPR75-ASB3
ASB3
|
G protein-coupled receptor 75 GPR75-ASB3 readthrough Ankyrin repeat and SOCS box protein 3 |
| chr1_-_52456352 | 0.03 |
ENST00000371655.3
|
RAB3B
|
RAB3B, member RAS oncogene family |
| chr2_+_7057523 | 0.03 |
ENST00000320892.6
|
RNF144A
|
ring finger protein 144A |
| chr10_-_97321112 | 0.03 |
ENST00000607232.1
ENST00000371227.4 ENST00000371249.2 ENST00000371247.2 ENST00000371246.2 ENST00000393949.1 ENST00000353505.5 ENST00000347291.4 |
SORBS1
|
sorbin and SH3 domain containing 1 |
| chr20_+_6748311 | 0.03 |
ENST00000378827.4
|
BMP2
|
bone morphogenetic protein 2 |
| chr3_-_141868357 | 0.02 |
ENST00000489671.1
ENST00000475734.1 ENST00000467072.1 ENST00000499676.2 |
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr1_-_154842741 | 0.02 |
ENST00000271915.4
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
| chr4_+_142557717 | 0.02 |
ENST00000320650.4
ENST00000296545.7 |
IL15
|
interleukin 15 |
| chr1_+_64239657 | 0.02 |
ENST00000371080.1
ENST00000371079.1 |
ROR1
|
receptor tyrosine kinase-like orphan receptor 1 |
| chr3_+_169684553 | 0.02 |
ENST00000337002.4
ENST00000480708.1 |
SEC62
|
SEC62 homolog (S. cerevisiae) |
| chr17_+_28804380 | 0.02 |
ENST00000225724.5
ENST00000451249.2 ENST00000467337.2 ENST00000581721.1 ENST00000414833.2 |
GOSR1
|
golgi SNAP receptor complex member 1 |
| chr8_-_103425047 | 0.02 |
ENST00000520539.1
|
UBR5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chr10_+_133753533 | 0.02 |
ENST00000422256.2
|
PPP2R2D
|
protein phosphatase 2, regulatory subunit B, delta |
| chr2_+_238707388 | 0.02 |
ENST00000316997.4
|
RBM44
|
RNA binding motif protein 44 |
| chr4_-_41216619 | 0.02 |
ENST00000508676.1
ENST00000506352.1 ENST00000295974.8 |
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr6_-_99395787 | 0.02 |
ENST00000369244.2
ENST00000229971.1 |
FBXL4
|
F-box and leucine-rich repeat protein 4 |
| chr7_+_155089486 | 0.02 |
ENST00000340368.4
ENST00000344756.4 ENST00000425172.1 ENST00000342407.5 |
INSIG1
|
insulin induced gene 1 |
| chr20_+_2795609 | 0.02 |
ENST00000554164.1
ENST00000380593.4 |
TMEM239
TMEM239
|
transmembrane protein 239 CDNA FLJ26142 fis, clone TST04526; Transmembrane protein 239; Uncharacterized protein |
| chr15_+_44719394 | 0.02 |
ENST00000260327.4
ENST00000396780.1 |
CTDSPL2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr18_-_5296001 | 0.02 |
ENST00000357006.4
|
ZBTB14
|
zinc finger and BTB domain containing 14 |
| chr21_+_40177143 | 0.02 |
ENST00000360214.3
|
ETS2
|
v-ets avian erythroblastosis virus E26 oncogene homolog 2 |
| chr2_+_46769798 | 0.02 |
ENST00000238738.4
|
RHOQ
|
ras homolog family member Q |
| chr2_-_175351744 | 0.02 |
ENST00000295500.4
ENST00000392552.2 ENST00000392551.2 |
GPR155
|
G protein-coupled receptor 155 |
| chr7_-_72936531 | 0.02 |
ENST00000339594.4
|
BAZ1B
|
bromodomain adjacent to zinc finger domain, 1B |
| chr1_+_117910047 | 0.02 |
ENST00000356554.3
|
MAN1A2
|
mannosidase, alpha, class 1A, member 2 |
| chr19_-_40030861 | 0.02 |
ENST00000390658.2
|
EID2
|
EP300 interacting inhibitor of differentiation 2 |
| chr11_-_106889250 | 0.02 |
ENST00000526355.2
|
GUCY1A2
|
guanylate cyclase 1, soluble, alpha 2 |
| chrX_-_119603138 | 0.02 |
ENST00000200639.4
ENST00000371335.4 ENST00000538785.1 ENST00000434600.2 |
LAMP2
|
lysosomal-associated membrane protein 2 |
| chr2_+_26256938 | 0.02 |
ENST00000264710.4
|
RAB10
|
RAB10, member RAS oncogene family |
| chr4_+_113152881 | 0.02 |
ENST00000274000.5
|
AP1AR
|
adaptor-related protein complex 1 associated regulatory protein |
| chr1_+_161736072 | 0.02 |
ENST00000367942.3
|
ATF6
|
activating transcription factor 6 |
| chr3_+_63638372 | 0.02 |
ENST00000496807.1
|
SNTN
|
sentan, cilia apical structure protein |
| chr9_-_13279589 | 0.02 |
ENST00000319217.7
|
MPDZ
|
multiple PDZ domain protein |
| chr2_+_149402553 | 0.02 |
ENST00000258484.6
ENST00000409654.1 |
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr12_-_82752565 | 0.02 |
ENST00000256151.7
|
CCDC59
|
coiled-coil domain containing 59 |
| chr6_+_116892530 | 0.02 |
ENST00000466444.2
ENST00000368590.5 ENST00000392526.1 |
RWDD1
|
RWD domain containing 1 |
| chr6_+_74405501 | 0.02 |
ENST00000437994.2
ENST00000422508.2 |
CD109
|
CD109 molecule |
| chr5_+_138629417 | 0.02 |
ENST00000510056.1
ENST00000511249.1 ENST00000503811.1 ENST00000511378.1 |
MATR3
|
matrin 3 |
| chr22_-_45636650 | 0.02 |
ENST00000336156.5
|
KIAA0930
|
KIAA0930 |
| chr1_+_14925173 | 0.02 |
ENST00000376030.2
ENST00000503743.1 ENST00000422387.2 |
KAZN
|
kazrin, periplakin interacting protein |
| chr9_+_15553055 | 0.02 |
ENST00000380701.3
|
CCDC171
|
coiled-coil domain containing 171 |
| chr7_-_35734730 | 0.02 |
ENST00000396081.1
ENST00000311350.3 |
HERPUD2
|
HERPUD family member 2 |
| chr4_+_148402069 | 0.02 |
ENST00000358556.4
ENST00000339690.5 ENST00000511804.1 ENST00000324300.5 |
EDNRA
|
endothelin receptor type A |
| chr2_+_192542850 | 0.02 |
ENST00000410026.2
|
NABP1
|
nucleic acid binding protein 1 |
| chr4_+_40751914 | 0.02 |
ENST00000381782.2
ENST00000316607.5 |
NSUN7
|
NOP2/Sun domain family, member 7 |
| chr3_-_57678772 | 0.02 |
ENST00000311128.5
|
DENND6A
|
DENN/MADD domain containing 6A |
| chr10_-_91295304 | 0.02 |
ENST00000341233.4
ENST00000371790.4 |
SLC16A12
|
solute carrier family 16, member 12 |
| chr5_+_76506706 | 0.02 |
ENST00000340978.3
ENST00000346042.3 ENST00000264917.5 ENST00000342343.4 ENST00000333194.4 |
PDE8B
|
phosphodiesterase 8B |
| chr5_-_148930960 | 0.02 |
ENST00000261798.5
ENST00000377843.2 |
CSNK1A1
|
casein kinase 1, alpha 1 |
| chr16_+_67596310 | 0.02 |
ENST00000264010.4
ENST00000401394.1 |
CTCF
|
CCCTC-binding factor (zinc finger protein) |
| chr17_+_30264014 | 0.02 |
ENST00000322652.5
ENST00000580398.1 |
SUZ12
|
SUZ12 polycomb repressive complex 2 subunit |
| chr9_-_117568365 | 0.02 |
ENST00000374045.4
|
TNFSF15
|
tumor necrosis factor (ligand) superfamily, member 15 |
| chr4_+_174089904 | 0.02 |
ENST00000265000.4
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) |
| chr10_+_20105157 | 0.02 |
ENST00000377242.3
ENST00000377252.4 |
PLXDC2
|
plexin domain containing 2 |
| chr15_+_74833518 | 0.02 |
ENST00000346246.5
|
ARID3B
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr17_-_57784755 | 0.02 |
ENST00000537860.1
ENST00000393038.2 ENST00000409433.2 |
PTRH2
|
peptidyl-tRNA hydrolase 2 |
| chr3_+_142720366 | 0.02 |
ENST00000493782.1
ENST00000397933.2 ENST00000473835.2 ENST00000493598.2 |
U2SURP
|
U2 snRNP-associated SURP domain containing |
| chr1_-_46152174 | 0.02 |
ENST00000290795.3
ENST00000355105.3 |
GPBP1L1
|
GC-rich promoter binding protein 1-like 1 |
| chr10_+_114206956 | 0.02 |
ENST00000432306.1
ENST00000393077.2 |
VTI1A
|
vesicle transport through interaction with t-SNAREs 1A |
| chr5_-_121413974 | 0.02 |
ENST00000231004.4
|
LOX
|
lysyl oxidase |
| chr11_-_76091986 | 0.02 |
ENST00000260045.3
|
PRKRIR
|
protein-kinase, interferon-inducible double stranded RNA dependent inhibitor, repressor of (P58 repressor) |
| chr4_+_146402925 | 0.02 |
ENST00000302085.4
|
SMAD1
|
SMAD family member 1 |
| chr10_+_79793518 | 0.02 |
ENST00000440692.1
ENST00000435275.1 ENST00000372360.3 ENST00000360830.4 |
RPS24
|
ribosomal protein S24 |
| chr5_+_56111361 | 0.02 |
ENST00000399503.3
|
MAP3K1
|
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
| chr12_+_124118366 | 0.02 |
ENST00000539994.1
ENST00000538845.1 ENST00000228955.7 ENST00000543341.2 ENST00000536375.1 |
GTF2H3
|
general transcription factor IIH, polypeptide 3, 34kDa |
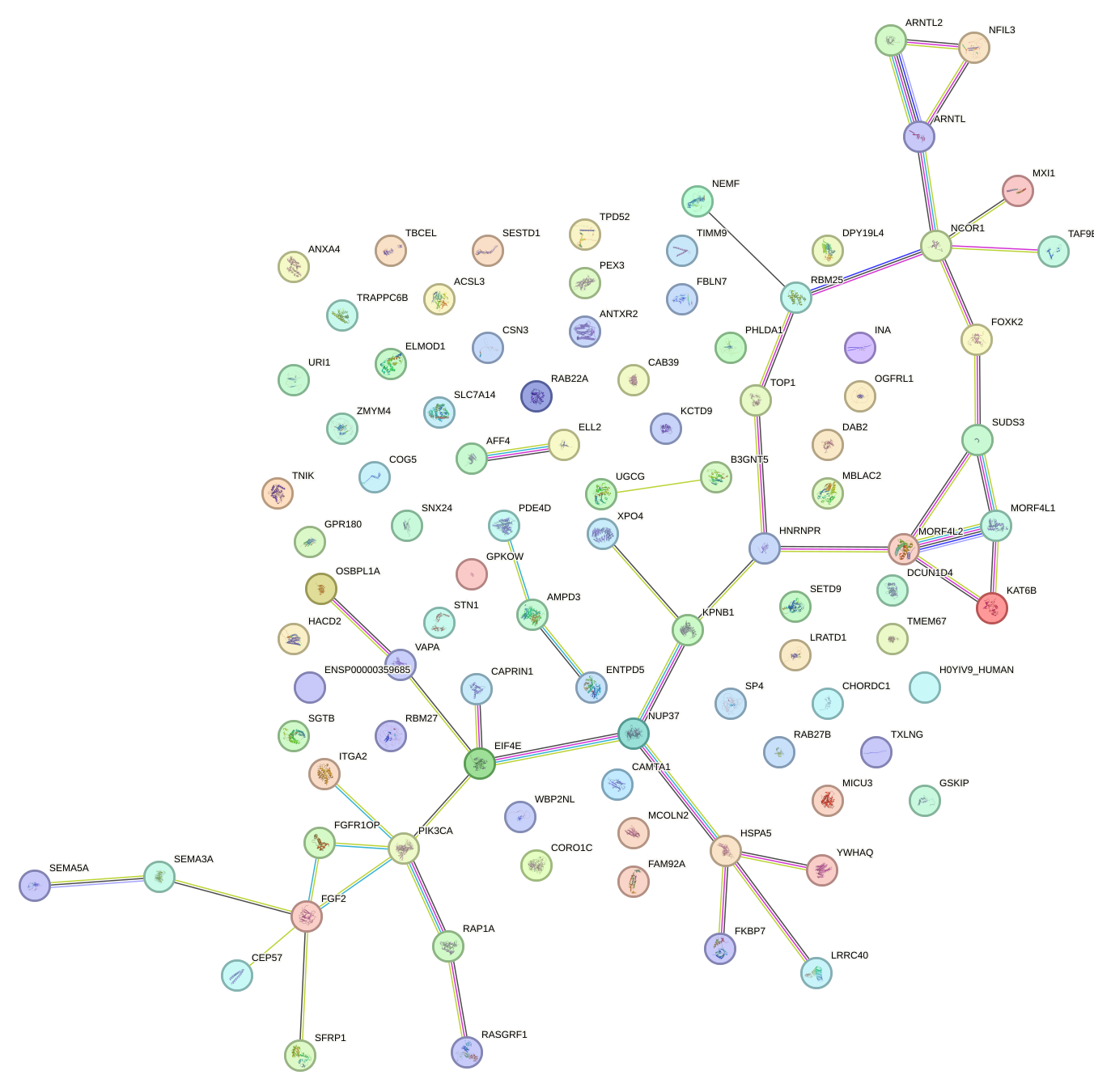
Network of associatons between targets according to the STRING database.
First level regulatory network of UGAAAUG
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.0 | 0.1 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 0.0 | 0.1 | GO:0045210 | FasL biosynthetic process(GO:0045210) |
| 0.0 | 0.1 | GO:0006679 | glucosylceramide biosynthetic process(GO:0006679) |
| 0.0 | 0.1 | GO:0042660 | positive regulation of cell fate specification(GO:0042660) |
| 0.0 | 0.1 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 0.0 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.0 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.0 | 0.0 | GO:2000681 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.0 | 0.1 | GO:2000301 | response to antineoplastic agent(GO:0097327) negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.0 | GO:2001247 | positive regulation of phosphatidylcholine biosynthetic process(GO:2001247) |
| 0.0 | 0.1 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.0 | 0.0 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.0 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.0 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.1 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.0 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.0 | GO:0015432 | bile acid-exporting ATPase activity(GO:0015432) |