Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
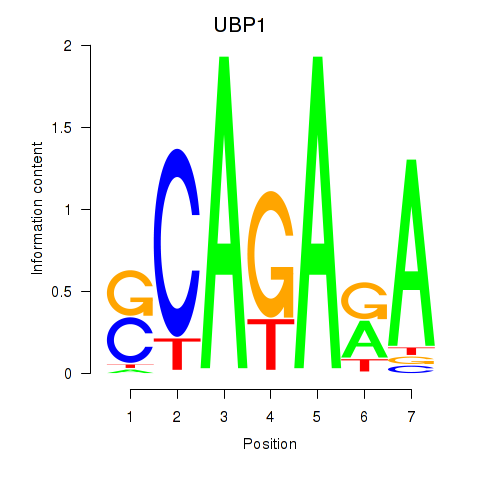
Results for UBP1
Z-value: 0.45
Transcription factors associated with UBP1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
UBP1
|
ENSG00000153560.7 | upstream binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| UBP1 | hg19_v2_chr3_-_33482002_33482116 | 0.29 | 7.1e-01 | Click! |
Activity profile of UBP1 motif
Sorted Z-values of UBP1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_35685452 | 0.16 |
ENST00000607559.1
|
TPM2
|
tropomyosin 2 (beta) |
| chr2_-_163175133 | 0.14 |
ENST00000421365.2
ENST00000263642.2 |
IFIH1
|
interferon induced with helicase C domain 1 |
| chr7_+_142829162 | 0.12 |
ENST00000291009.3
|
PIP
|
prolactin-induced protein |
| chr17_-_5138099 | 0.11 |
ENST00000571800.1
ENST00000574081.1 ENST00000399600.4 ENST00000574297.1 |
SCIMP
|
SLP adaptor and CSK interacting membrane protein |
| chr22_+_22676808 | 0.11 |
ENST00000390290.2
|
IGLV1-51
|
immunoglobulin lambda variable 1-51 |
| chr9_-_14322319 | 0.10 |
ENST00000606230.1
|
NFIB
|
nuclear factor I/B |
| chr2_+_162087577 | 0.09 |
ENST00000439442.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr11_-_134123142 | 0.09 |
ENST00000392595.2
ENST00000341541.3 ENST00000352327.5 ENST00000392594.3 |
THYN1
|
thymocyte nuclear protein 1 |
| chr5_+_122847908 | 0.09 |
ENST00000511130.2
ENST00000512718.3 |
CSNK1G3
|
casein kinase 1, gamma 3 |
| chr11_-_11374904 | 0.09 |
ENST00000528848.2
|
CSNK2A3
|
casein kinase 2, alpha 3 polypeptide |
| chr2_+_170550944 | 0.09 |
ENST00000359744.3
ENST00000438838.1 ENST00000438710.1 ENST00000449906.1 ENST00000498202.2 ENST00000272797.4 |
PHOSPHO2
KLHL23
|
phosphatase, orphan 2 kelch-like family member 23 |
| chr1_-_185126037 | 0.09 |
ENST00000367506.5
ENST00000367504.3 |
TRMT1L
|
tRNA methyltransferase 1 homolog (S. cerevisiae)-like |
| chr19_+_38880695 | 0.08 |
ENST00000587947.1
ENST00000338502.4 |
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr5_+_95998070 | 0.08 |
ENST00000421689.2
ENST00000510756.1 ENST00000512620.1 |
CAST
|
calpastatin |
| chr9_+_15422702 | 0.08 |
ENST00000380821.3
ENST00000421710.1 |
SNAPC3
|
small nuclear RNA activating complex, polypeptide 3, 50kDa |
| chr15_-_77712429 | 0.08 |
ENST00000564328.1
ENST00000558305.1 |
PEAK1
|
pseudopodium-enriched atypical kinase 1 |
| chr9_-_6015607 | 0.07 |
ENST00000259569.5
|
RANBP6
|
RAN binding protein 6 |
| chr1_+_63989004 | 0.07 |
ENST00000371088.4
|
EFCAB7
|
EF-hand calcium binding domain 7 |
| chr19_-_51875523 | 0.07 |
ENST00000593572.1
ENST00000595157.1 |
NKG7
|
natural killer cell group 7 sequence |
| chr12_+_6898674 | 0.07 |
ENST00000541982.1
ENST00000539492.1 |
CD4
|
CD4 molecule |
| chr19_-_58459039 | 0.06 |
ENST00000282308.3
ENST00000598928.1 |
ZNF256
|
zinc finger protein 256 |
| chr5_-_93447333 | 0.06 |
ENST00000395965.3
ENST00000505869.1 ENST00000509163.1 |
FAM172A
|
family with sequence similarity 172, member A |
| chr5_+_122847781 | 0.06 |
ENST00000395412.1
ENST00000395411.1 ENST00000345990.4 |
CSNK1G3
|
casein kinase 1, gamma 3 |
| chr2_-_170550842 | 0.06 |
ENST00000421028.1
|
CCDC173
|
coiled-coil domain containing 173 |
| chr7_+_101460882 | 0.06 |
ENST00000292535.7
ENST00000549414.2 ENST00000550008.2 ENST00000546411.2 ENST00000556210.1 |
CUX1
|
cut-like homeobox 1 |
| chr12_+_123717458 | 0.06 |
ENST00000253233.1
|
C12orf65
|
chromosome 12 open reading frame 65 |
| chr6_+_142623758 | 0.06 |
ENST00000541199.1
ENST00000435011.2 |
GPR126
|
G protein-coupled receptor 126 |
| chr4_-_100815525 | 0.06 |
ENST00000226522.8
ENST00000499666.2 |
LAMTOR3
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 3 |
| chr12_-_123717643 | 0.05 |
ENST00000541437.1
ENST00000606320.1 |
MPHOSPH9
|
M-phase phosphoprotein 9 |
| chr7_+_105172612 | 0.05 |
ENST00000493041.1
|
RINT1
|
RAD50 interactor 1 |
| chr19_-_59066327 | 0.05 |
ENST00000596708.1
ENST00000601220.1 ENST00000597848.1 |
CHMP2A
|
charged multivesicular body protein 2A |
| chr1_+_48688357 | 0.05 |
ENST00000533824.1
ENST00000438567.2 ENST00000236495.5 ENST00000420136.2 |
SLC5A9
|
solute carrier family 5 (sodium/sugar cotransporter), member 9 |
| chr11_+_7506713 | 0.05 |
ENST00000329293.3
ENST00000534244.1 |
OLFML1
|
olfactomedin-like 1 |
| chr5_-_41510656 | 0.05 |
ENST00000377801.3
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr1_-_150208498 | 0.05 |
ENST00000314136.8
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr1_-_150669604 | 0.04 |
ENST00000427665.1
ENST00000540514.1 |
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr6_+_31555045 | 0.04 |
ENST00000396101.3
ENST00000490742.1 |
LST1
|
leukocyte specific transcript 1 |
| chrX_+_74493896 | 0.04 |
ENST00000373383.4
ENST00000373379.1 |
UPRT
|
uracil phosphoribosyltransferase (FUR1) homolog (S. cerevisiae) |
| chr10_-_75676400 | 0.04 |
ENST00000412307.2
|
C10orf55
|
chromosome 10 open reading frame 55 |
| chr12_-_123717711 | 0.04 |
ENST00000537854.1
|
MPHOSPH9
|
M-phase phosphoprotein 9 |
| chr22_+_38071615 | 0.04 |
ENST00000215909.5
|
LGALS1
|
lectin, galactoside-binding, soluble, 1 |
| chr13_-_23949671 | 0.04 |
ENST00000402364.1
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr11_+_124824000 | 0.04 |
ENST00000529051.1
ENST00000344762.5 |
CCDC15
|
coiled-coil domain containing 15 |
| chr6_+_6588316 | 0.04 |
ENST00000379953.2
|
LY86
|
lymphocyte antigen 86 |
| chr14_-_77542485 | 0.04 |
ENST00000556781.1
ENST00000557526.1 ENST00000555512.1 |
RP11-7F17.3
|
RP11-7F17.3 |
| chr6_-_154567984 | 0.04 |
ENST00000517438.1
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1 |
| chr3_+_29323043 | 0.03 |
ENST00000452462.1
ENST00000456853.1 |
RBMS3
|
RNA binding motif, single stranded interacting protein 3 |
| chr15_-_42343388 | 0.03 |
ENST00000399518.3
|
PLA2G4E
|
phospholipase A2, group IVE |
| chr17_+_48450575 | 0.03 |
ENST00000338165.4
ENST00000393271.2 ENST00000511519.2 |
EME1
|
essential meiotic structure-specific endonuclease 1 |
| chrX_+_38420623 | 0.03 |
ENST00000378482.2
|
TSPAN7
|
tetraspanin 7 |
| chrY_+_2803322 | 0.03 |
ENST00000383052.1
ENST00000155093.3 ENST00000449237.1 ENST00000443793.1 |
ZFY
|
zinc finger protein, Y-linked |
| chr17_-_79304150 | 0.03 |
ENST00000574093.1
|
TMEM105
|
transmembrane protein 105 |
| chr8_+_28747884 | 0.03 |
ENST00000287701.10
ENST00000444075.1 ENST00000403668.2 ENST00000519662.1 ENST00000558662.1 ENST00000523613.1 ENST00000560599.1 ENST00000397358.3 |
HMBOX1
|
homeobox containing 1 |
| chr15_-_77712477 | 0.03 |
ENST00000560626.2
|
PEAK1
|
pseudopodium-enriched atypical kinase 1 |
| chr8_-_28747717 | 0.03 |
ENST00000416984.2
|
INTS9
|
integrator complex subunit 9 |
| chr1_-_150208412 | 0.03 |
ENST00000532744.1
ENST00000369114.5 ENST00000369115.2 ENST00000369116.4 |
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr15_-_101835110 | 0.03 |
ENST00000560496.1
|
SNRPA1
|
small nuclear ribonucleoprotein polypeptide A' |
| chr1_-_150208363 | 0.03 |
ENST00000436748.2
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr4_+_55096489 | 0.03 |
ENST00000504461.1
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr1_-_202129105 | 0.03 |
ENST00000367279.4
|
PTPN7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr7_+_150706010 | 0.02 |
ENST00000475017.1
|
NOS3
|
nitric oxide synthase 3 (endothelial cell) |
| chr1_-_202129704 | 0.02 |
ENST00000476061.1
ENST00000544762.1 ENST00000467283.1 ENST00000464870.1 ENST00000435759.2 ENST00000486116.1 ENST00000543735.1 ENST00000308986.5 ENST00000477625.1 |
PTPN7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr7_+_29874341 | 0.02 |
ENST00000409290.1
ENST00000242140.5 |
WIPF3
|
WAS/WASL interacting protein family, member 3 |
| chr1_-_156217875 | 0.02 |
ENST00000292291.5
|
PAQR6
|
progestin and adipoQ receptor family member VI |
| chr9_+_108463234 | 0.02 |
ENST00000374688.1
|
TMEM38B
|
transmembrane protein 38B |
| chr11_+_128761220 | 0.02 |
ENST00000529694.1
|
KCNJ5
|
potassium inwardly-rectifying channel, subfamily J, member 5 |
| chr3_+_111718036 | 0.02 |
ENST00000455401.2
|
TAGLN3
|
transgelin 3 |
| chr1_-_150208291 | 0.02 |
ENST00000533654.1
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr5_+_34656331 | 0.02 |
ENST00000265109.3
|
RAI14
|
retinoic acid induced 14 |
| chr1_-_182369751 | 0.02 |
ENST00000367565.1
|
TEDDM1
|
transmembrane epididymal protein 1 |
| chr14_-_23395623 | 0.02 |
ENST00000556043.1
|
PRMT5
|
protein arginine methyltransferase 5 |
| chr15_+_41136734 | 0.02 |
ENST00000568580.1
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr1_-_168698433 | 0.01 |
ENST00000367817.3
|
DPT
|
dermatopontin |
| chr9_+_109685630 | 0.01 |
ENST00000451160.2
|
RP11-508N12.4
|
Uncharacterized protein |
| chr9_+_130853715 | 0.01 |
ENST00000373066.5
ENST00000432073.2 |
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25 |
| chr3_+_111260980 | 0.01 |
ENST00000438817.2
|
CD96
|
CD96 molecule |
| chr3_-_56502375 | 0.01 |
ENST00000288221.6
|
ERC2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr1_-_144995002 | 0.01 |
ENST00000369356.4
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chrX_+_139791917 | 0.01 |
ENST00000607004.1
ENST00000370535.3 |
LINC00632
|
long intergenic non-protein coding RNA 632 |
| chr1_-_114302086 | 0.01 |
ENST00000369604.1
ENST00000357783.2 |
PHTF1
|
putative homeodomain transcription factor 1 |
| chr7_+_132937820 | 0.01 |
ENST00000393161.2
ENST00000253861.4 |
EXOC4
|
exocyst complex component 4 |
| chr10_-_27443294 | 0.01 |
ENST00000396296.3
ENST00000375972.3 ENST00000376016.3 ENST00000491542.2 |
YME1L1
|
YME1-like 1 ATPase |
| chr4_-_47839966 | 0.01 |
ENST00000273857.4
ENST00000505909.1 ENST00000502252.1 |
CORIN
|
corin, serine peptidase |
| chr15_+_34638066 | 0.01 |
ENST00000333756.4
|
NUTM1
|
NUT midline carcinoma, family member 1 |
| chr11_-_14521379 | 0.01 |
ENST00000249923.3
ENST00000529866.1 ENST00000439561.2 ENST00000534771.1 |
COPB1
|
coatomer protein complex, subunit beta 1 |
| chr3_+_111260954 | 0.01 |
ENST00000283285.5
|
CD96
|
CD96 molecule |
| chrX_+_115567767 | 0.01 |
ENST00000371900.4
|
SLC6A14
|
solute carrier family 6 (amino acid transporter), member 14 |
| chr1_-_144995074 | 0.01 |
ENST00000534536.1
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr1_-_152332480 | 0.01 |
ENST00000388718.5
|
FLG2
|
filaggrin family member 2 |
| chr9_+_112852477 | 0.01 |
ENST00000480388.1
|
AKAP2
|
A kinase (PRKA) anchor protein 2 |
| chr12_+_101188547 | 0.00 |
ENST00000546991.1
ENST00000392979.3 |
ANO4
|
anoctamin 4 |
| chr14_-_92413727 | 0.00 |
ENST00000267620.10
|
FBLN5
|
fibulin 5 |
| chrX_-_10851762 | 0.00 |
ENST00000380785.1
ENST00000380787.1 |
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr12_-_47473425 | 0.00 |
ENST00000550413.1
|
AMIGO2
|
adhesion molecule with Ig-like domain 2 |
| chr2_+_17721920 | 0.00 |
ENST00000295156.4
|
VSNL1
|
visinin-like 1 |
| chr2_+_113033164 | 0.00 |
ENST00000409871.1
ENST00000343936.4 |
ZC3H6
|
zinc finger CCCH-type containing 6 |
| chr3_+_111260856 | 0.00 |
ENST00000352690.4
|
CD96
|
CD96 molecule |
| chr5_+_127039075 | 0.00 |
ENST00000514853.2
|
CTC-228N24.1
|
CTC-228N24.1 |
| chr8_-_28747424 | 0.00 |
ENST00000523436.1
ENST00000397363.4 ENST00000521777.1 ENST00000520184.1 ENST00000521022.1 |
INTS9
|
integrator complex subunit 9 |
Network of associatons between targets according to the STRING database.
First level regulatory network of UBP1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.0 | 0.1 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.0 | 0.1 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.1 | GO:0035723 | interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-15(GO:0071350) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070939 | Dsl1p complex(GO:0070939) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |