Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
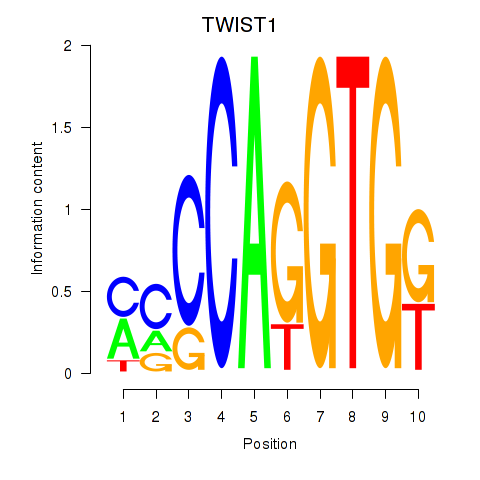
Results for TWIST1_SNAI1
Z-value: 0.54
Transcription factors associated with TWIST1_SNAI1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TWIST1
|
ENSG00000122691.8 | twist family bHLH transcription factor 1 |
|
SNAI1
|
ENSG00000124216.3 | snail family transcriptional repressor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SNAI1 | hg19_v2_chr20_+_48599506_48599536 | 0.99 | 1.1e-02 | Click! |
| TWIST1 | hg19_v2_chr7_-_19157248_19157295 | -0.65 | 3.5e-01 | Click! |
Activity profile of TWIST1_SNAI1 motif
Sorted Z-values of TWIST1_SNAI1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_5567842 | 0.38 |
ENST00000587632.1
|
TINCR
|
tissue differentiation-inducing non-protein coding RNA |
| chr2_+_220306238 | 0.35 |
ENST00000435853.1
|
SPEG
|
SPEG complex locus |
| chr9_-_140142222 | 0.32 |
ENST00000344774.4
ENST00000388932.2 |
FAM166A
|
family with sequence similarity 166, member A |
| chr19_-_56109119 | 0.29 |
ENST00000587678.1
|
FIZ1
|
FLT3-interacting zinc finger 1 |
| chr19_-_40730820 | 0.27 |
ENST00000513948.1
|
CNTD2
|
cyclin N-terminal domain containing 2 |
| chr1_-_1850697 | 0.25 |
ENST00000378598.4
ENST00000416272.1 ENST00000310991.3 |
TMEM52
|
transmembrane protein 52 |
| chr19_-_48673465 | 0.24 |
ENST00000598938.1
|
LIG1
|
ligase I, DNA, ATP-dependent |
| chr3_+_49059038 | 0.23 |
ENST00000451378.2
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr12_+_8849773 | 0.23 |
ENST00000541044.1
|
RIMKLB
|
ribosomal modification protein rimK-like family member B |
| chr1_-_95285652 | 0.23 |
ENST00000442418.1
|
LINC01057
|
long intergenic non-protein coding RNA 1057 |
| chr17_-_42452063 | 0.22 |
ENST00000588098.1
|
ITGA2B
|
integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41) |
| chr15_+_31658349 | 0.21 |
ENST00000558844.1
|
KLF13
|
Kruppel-like factor 13 |
| chr19_-_7939319 | 0.21 |
ENST00000539422.1
|
CTD-3193O13.9
|
Protein FLJ22184 |
| chr8_-_17270809 | 0.19 |
ENST00000180173.5
ENST00000521857.1 |
MTMR7
|
myotubularin related protein 7 |
| chr19_-_49843539 | 0.17 |
ENST00000602554.1
ENST00000358234.4 |
CTC-301O7.4
|
CTC-301O7.4 |
| chr19_-_51289374 | 0.17 |
ENST00000563228.1
|
CTD-2568A17.1
|
CTD-2568A17.1 |
| chr11_+_117049910 | 0.17 |
ENST00000431081.2
ENST00000524842.1 |
SIDT2
|
SID1 transmembrane family, member 2 |
| chr1_-_22215192 | 0.16 |
ENST00000374673.3
|
HSPG2
|
heparan sulfate proteoglycan 2 |
| chr1_+_35247859 | 0.14 |
ENST00000373362.3
|
GJB3
|
gap junction protein, beta 3, 31kDa |
| chr6_-_3227877 | 0.14 |
ENST00000259818.7
|
TUBB2B
|
tubulin, beta 2B class IIb |
| chr20_-_62493217 | 0.13 |
ENST00000601296.1
|
C20ORF135
|
C20ORF135 |
| chr19_+_39759154 | 0.13 |
ENST00000331982.5
|
IFNL2
|
interferon, lambda 2 |
| chr22_+_31489344 | 0.13 |
ENST00000404574.1
|
SMTN
|
smoothelin |
| chr17_+_52978156 | 0.13 |
ENST00000348161.4
|
TOM1L1
|
target of myb1 (chicken)-like 1 |
| chr17_+_77030267 | 0.13 |
ENST00000581774.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr12_+_54384370 | 0.13 |
ENST00000504315.1
|
HOXC6
|
homeobox C6 |
| chr3_-_120169828 | 0.13 |
ENST00000424703.2
ENST00000469005.1 |
FSTL1
|
follistatin-like 1 |
| chr22_+_50312316 | 0.13 |
ENST00000328268.4
|
CRELD2
|
cysteine-rich with EGF-like domains 2 |
| chr12_+_52450298 | 0.12 |
ENST00000550582.2
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr20_+_48599506 | 0.12 |
ENST00000244050.2
|
SNAI1
|
snail family zinc finger 1 |
| chr19_-_49565254 | 0.12 |
ENST00000593537.1
|
NTF4
|
neurotrophin 4 |
| chr1_+_179712298 | 0.12 |
ENST00000341785.4
|
FAM163A
|
family with sequence similarity 163, member A |
| chr17_-_40264692 | 0.12 |
ENST00000591220.1
ENST00000251642.3 |
DHX58
|
DEXH (Asp-Glu-X-His) box polypeptide 58 |
| chr2_+_27665289 | 0.11 |
ENST00000407293.1
|
KRTCAP3
|
keratinocyte associated protein 3 |
| chr11_+_117049854 | 0.11 |
ENST00000278951.7
|
SIDT2
|
SID1 transmembrane family, member 2 |
| chr11_-_117667806 | 0.11 |
ENST00000527706.1
ENST00000321322.6 |
DSCAML1
|
Down syndrome cell adhesion molecule like 1 |
| chr11_-_88070920 | 0.11 |
ENST00000524463.1
ENST00000227266.5 |
CTSC
|
cathepsin C |
| chr13_+_114239588 | 0.11 |
ENST00000544902.1
ENST00000408980.2 ENST00000453989.1 |
TFDP1
|
transcription factor Dp-1 |
| chr17_+_39421591 | 0.11 |
ENST00000391355.1
|
KRTAP9-6
|
keratin associated protein 9-6 |
| chr19_-_10420459 | 0.11 |
ENST00000403352.1
ENST00000403903.3 |
ZGLP1
|
zinc finger, GATA-like protein 1 |
| chr17_-_18945798 | 0.11 |
ENST00000395635.1
|
GRAP
|
GRB2-related adaptor protein |
| chr4_+_4387983 | 0.11 |
ENST00000397958.1
|
NSG1
|
Homo sapiens neuron specific gene family member 1 (NSG1), transcript variant 3, mRNA. |
| chr3_+_72201910 | 0.11 |
ENST00000469178.1
ENST00000485404.1 |
LINC00870
|
long intergenic non-protein coding RNA 870 |
| chr12_+_27091426 | 0.11 |
ENST00000546072.1
ENST00000327214.5 |
FGFR1OP2
|
FGFR1 oncogene partner 2 |
| chr22_+_24820341 | 0.11 |
ENST00000464977.1
ENST00000444262.2 |
ADORA2A
|
adenosine A2a receptor |
| chr11_+_32605350 | 0.11 |
ENST00000531120.1
ENST00000524896.1 ENST00000323213.5 |
EIF3M
|
eukaryotic translation initiation factor 3, subunit M |
| chr1_+_3385085 | 0.11 |
ENST00000445297.1
|
ARHGEF16
|
Rho guanine nucleotide exchange factor (GEF) 16 |
| chr8_+_145065705 | 0.11 |
ENST00000533044.1
|
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr19_+_54926621 | 0.11 |
ENST00000376530.3
ENST00000445095.1 ENST00000391739.3 ENST00000376531.3 |
TTYH1
|
tweety family member 1 |
| chr20_-_62103862 | 0.11 |
ENST00000344462.4
ENST00000357249.2 ENST00000359125.2 ENST00000360480.3 ENST00000370224.1 ENST00000344425.5 ENST00000354587.3 ENST00000359689.1 |
KCNQ2
|
potassium voltage-gated channel, KQT-like subfamily, member 2 |
| chrX_-_45710920 | 0.11 |
ENST00000456532.1
|
RP5-1158E12.3
|
RP5-1158E12.3 |
| chr11_-_118972575 | 0.11 |
ENST00000432443.2
|
DPAGT1
|
dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase) |
| chr11_-_407103 | 0.10 |
ENST00000526395.1
|
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr19_-_42806842 | 0.10 |
ENST00000596265.1
|
PAFAH1B3
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa) |
| chr17_-_73149921 | 0.10 |
ENST00000481647.1
ENST00000470924.1 |
HN1
|
hematological and neurological expressed 1 |
| chr16_+_25123148 | 0.10 |
ENST00000570981.1
|
LCMT1
|
leucine carboxyl methyltransferase 1 |
| chr11_-_57102947 | 0.10 |
ENST00000526696.1
|
SSRP1
|
structure specific recognition protein 1 |
| chr5_+_94890840 | 0.10 |
ENST00000504763.1
|
ARSK
|
arylsulfatase family, member K |
| chr7_-_127032114 | 0.10 |
ENST00000436992.1
|
ZNF800
|
zinc finger protein 800 |
| chr22_-_25801333 | 0.10 |
ENST00000444995.3
|
LRP5L
|
low density lipoprotein receptor-related protein 5-like |
| chr9_-_123638633 | 0.10 |
ENST00000456291.1
|
PHF19
|
PHD finger protein 19 |
| chr17_+_52978185 | 0.09 |
ENST00000572405.1
ENST00000572158.1 ENST00000540336.1 ENST00000572298.1 ENST00000536554.1 ENST00000575333.1 ENST00000570499.1 ENST00000572576.1 |
TOM1L1
|
target of myb1 (chicken)-like 1 |
| chr15_+_90611465 | 0.09 |
ENST00000559360.1
|
ZNF710
|
zinc finger protein 710 |
| chr13_+_31480328 | 0.09 |
ENST00000380482.4
|
MEDAG
|
mesenteric estrogen-dependent adipogenesis |
| chr11_+_45944190 | 0.09 |
ENST00000401752.1
ENST00000389968.3 ENST00000325468.5 ENST00000536139.1 |
GYLTL1B
|
glycosyltransferase-like 1B |
| chr12_-_110318130 | 0.09 |
ENST00000540772.1
|
GLTP
|
glycolipid transfer protein |
| chrX_+_115567767 | 0.09 |
ENST00000371900.4
|
SLC6A14
|
solute carrier family 6 (amino acid transporter), member 14 |
| chr9_-_136919256 | 0.09 |
ENST00000433041.1
|
BRD3
|
bromodomain containing 3 |
| chr19_+_1105110 | 0.09 |
ENST00000587648.1
|
GPX4
|
glutathione peroxidase 4 |
| chr8_+_67687413 | 0.09 |
ENST00000521960.1
ENST00000522398.1 ENST00000522629.1 ENST00000520976.1 ENST00000396596.1 |
SGK3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr19_+_35532612 | 0.09 |
ENST00000600390.1
ENST00000597419.1 |
HPN
|
hepsin |
| chrX_-_45060135 | 0.09 |
ENST00000398000.2
ENST00000377934.4 |
CXorf36
|
chromosome X open reading frame 36 |
| chr12_-_49259643 | 0.09 |
ENST00000309739.5
|
RND1
|
Rho family GTPase 1 |
| chr16_-_29757272 | 0.09 |
ENST00000329410.3
|
C16orf54
|
chromosome 16 open reading frame 54 |
| chr19_-_3600549 | 0.09 |
ENST00000589966.1
|
TBXA2R
|
thromboxane A2 receptor |
| chr12_+_65174519 | 0.09 |
ENST00000229088.6
|
TBC1D30
|
TBC1 domain family, member 30 |
| chr8_+_145065521 | 0.09 |
ENST00000534791.1
|
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr15_+_80351977 | 0.09 |
ENST00000559157.1
ENST00000561012.1 ENST00000564367.1 ENST00000558494.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr4_+_144354644 | 0.09 |
ENST00000512843.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr19_-_13068012 | 0.09 |
ENST00000316939.1
|
GADD45GIP1
|
growth arrest and DNA-damage-inducible, gamma interacting protein 1 |
| chr12_-_124456598 | 0.09 |
ENST00000539761.1
ENST00000539551.1 |
CCDC92
|
coiled-coil domain containing 92 |
| chr1_+_95583479 | 0.08 |
ENST00000455656.1
ENST00000604534.1 |
TMEM56
RP11-57H12.6
|
transmembrane protein 56 TMEM56-RWDD3 readthrough |
| chr6_+_63921351 | 0.08 |
ENST00000370659.1
|
FKBP1C
|
FK506 binding protein 1C |
| chr16_+_23847339 | 0.08 |
ENST00000303531.7
|
PRKCB
|
protein kinase C, beta |
| chr8_-_81083731 | 0.08 |
ENST00000379096.5
|
TPD52
|
tumor protein D52 |
| chr2_+_232316906 | 0.08 |
ENST00000370380.2
|
AC017104.2
|
Uncharacterized protein |
| chr6_-_31138439 | 0.08 |
ENST00000259915.8
|
POU5F1
|
POU class 5 homeobox 1 |
| chr15_+_90728145 | 0.08 |
ENST00000561085.1
ENST00000379122.3 ENST00000332496.6 |
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr10_+_35416223 | 0.08 |
ENST00000489321.1
ENST00000427847.2 ENST00000345491.3 ENST00000395895.2 ENST00000374728.3 ENST00000487132.1 |
CREM
|
cAMP responsive element modulator |
| chr1_-_11107280 | 0.08 |
ENST00000400897.3
ENST00000400898.3 |
MASP2
|
mannan-binding lectin serine peptidase 2 |
| chr14_+_75894714 | 0.08 |
ENST00000559060.1
|
JDP2
|
Jun dimerization protein 2 |
| chr8_+_96037255 | 0.08 |
ENST00000286687.4
|
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr19_-_11545920 | 0.08 |
ENST00000356392.4
ENST00000591179.1 |
CCDC151
|
coiled-coil domain containing 151 |
| chr22_-_21905120 | 0.08 |
ENST00000331505.5
|
RIMBP3C
|
RIMS binding protein 3C |
| chr16_+_14980632 | 0.08 |
ENST00000565655.1
|
NOMO1
|
NODAL modulator 1 |
| chr22_+_46731676 | 0.08 |
ENST00000424260.2
|
TRMU
|
tRNA 5-methylaminomethyl-2-thiouridylate methyltransferase |
| chr6_-_31550192 | 0.08 |
ENST00000429299.2
ENST00000446745.2 |
LTB
|
lymphotoxin beta (TNF superfamily, member 3) |
| chr5_+_76506706 | 0.08 |
ENST00000340978.3
ENST00000346042.3 ENST00000264917.5 ENST00000342343.4 ENST00000333194.4 |
PDE8B
|
phosphodiesterase 8B |
| chr1_-_40367668 | 0.08 |
ENST00000397332.2
ENST00000429311.1 |
MYCL
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr1_-_55266865 | 0.08 |
ENST00000371274.4
|
TTC22
|
tetratricopeptide repeat domain 22 |
| chr3_-_124606502 | 0.08 |
ENST00000483168.1
|
ITGB5
|
integrin, beta 5 |
| chr18_+_3262415 | 0.08 |
ENST00000581193.1
ENST00000400175.5 |
MYL12B
|
myosin, light chain 12B, regulatory |
| chr11_-_2924720 | 0.08 |
ENST00000455942.2
|
SLC22A18AS
|
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr4_+_40198527 | 0.07 |
ENST00000381799.5
|
RHOH
|
ras homolog family member H |
| chr19_+_55996565 | 0.07 |
ENST00000587400.1
|
NAT14
|
N-acetyltransferase 14 (GCN5-related, putative) |
| chr1_-_156675564 | 0.07 |
ENST00000368220.1
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr2_+_219125714 | 0.07 |
ENST00000522678.1
ENST00000519574.1 ENST00000521462.1 |
GPBAR1
|
G protein-coupled bile acid receptor 1 |
| chr8_+_96037205 | 0.07 |
ENST00000396124.4
|
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr5_+_63461642 | 0.07 |
ENST00000296615.6
ENST00000381081.2 ENST00000389100.4 |
RNF180
|
ring finger protein 180 |
| chr16_+_318638 | 0.07 |
ENST00000412541.1
ENST00000435035.1 |
ARHGDIG
|
Rho GDP dissociation inhibitor (GDI) gamma |
| chr4_-_38806404 | 0.07 |
ENST00000308979.2
ENST00000505940.1 ENST00000515861.1 |
TLR1
|
toll-like receptor 1 |
| chr16_-_77468945 | 0.07 |
ENST00000282849.5
|
ADAMTS18
|
ADAM metallopeptidase with thrombospondin type 1 motif, 18 |
| chr2_+_219283815 | 0.07 |
ENST00000248444.5
ENST00000454069.1 ENST00000392114.2 |
VIL1
|
villin 1 |
| chr1_+_231376941 | 0.07 |
ENST00000436239.1
ENST00000366647.4 ENST00000366646.3 ENST00000416000.1 |
GNPAT
|
glyceronephosphate O-acyltransferase |
| chr10_+_75910960 | 0.07 |
ENST00000539909.1
ENST00000286621.2 |
ADK
|
adenosine kinase |
| chr8_+_356942 | 0.07 |
ENST00000276326.5
|
FBXO25
|
F-box protein 25 |
| chr14_+_105781102 | 0.07 |
ENST00000547217.1
|
PACS2
|
phosphofurin acidic cluster sorting protein 2 |
| chr15_+_74833518 | 0.07 |
ENST00000346246.5
|
ARID3B
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr1_+_17634689 | 0.07 |
ENST00000375453.1
ENST00000375448.4 |
PADI4
|
peptidyl arginine deiminase, type IV |
| chr1_+_153651078 | 0.07 |
ENST00000368680.3
|
NPR1
|
natriuretic peptide receptor A/guanylate cyclase A (atrionatriuretic peptide receptor A) |
| chr1_-_113498616 | 0.07 |
ENST00000433570.4
ENST00000538576.1 ENST00000458229.1 |
SLC16A1
|
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr8_+_144821557 | 0.07 |
ENST00000534398.1
|
FAM83H-AS1
|
FAM83H antisense RNA 1 (head to head) |
| chr16_-_2004683 | 0.07 |
ENST00000268661.7
|
RPL3L
|
ribosomal protein L3-like |
| chr12_-_121972556 | 0.07 |
ENST00000545022.1
|
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr18_+_3262954 | 0.07 |
ENST00000584539.1
|
MYL12B
|
myosin, light chain 12B, regulatory |
| chr22_+_50312379 | 0.07 |
ENST00000407217.3
ENST00000403427.3 |
CRELD2
|
cysteine-rich with EGF-like domains 2 |
| chr11_+_111750206 | 0.07 |
ENST00000530214.1
ENST00000530799.1 |
C11orf1
|
chromosome 11 open reading frame 1 |
| chr14_+_104029278 | 0.07 |
ENST00000472726.2
ENST00000409074.2 ENST00000440963.1 ENST00000556253.2 ENST00000247618.4 |
RP11-73M18.2
APOPT1
|
Kinesin light chain 1 apoptogenic 1, mitochondrial |
| chr11_-_65626797 | 0.07 |
ENST00000525451.2
|
CFL1
|
cofilin 1 (non-muscle) |
| chr2_-_242211359 | 0.07 |
ENST00000444092.1
|
HDLBP
|
high density lipoprotein binding protein |
| chr1_-_93426998 | 0.07 |
ENST00000370310.4
|
FAM69A
|
family with sequence similarity 69, member A |
| chr16_-_67493110 | 0.07 |
ENST00000602876.1
|
ATP6V0D1
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d1 |
| chr9_+_91003271 | 0.07 |
ENST00000375859.3
ENST00000541629.1 |
SPIN1
|
spindlin 1 |
| chr5_-_132200477 | 0.07 |
ENST00000296875.2
|
GDF9
|
growth differentiation factor 9 |
| chr17_-_1619491 | 0.07 |
ENST00000570416.1
ENST00000575626.1 ENST00000610106.1 ENST00000608198.1 ENST00000609442.1 ENST00000334146.3 ENST00000576489.1 ENST00000608245.1 ENST00000609398.1 ENST00000608913.1 ENST00000574016.1 ENST00000571091.1 ENST00000573127.1 ENST00000609990.1 ENST00000576749.1 |
MIR22HG
|
MIR22 host gene (non-protein coding) |
| chr4_+_89300158 | 0.07 |
ENST00000502870.1
|
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr15_+_69591235 | 0.07 |
ENST00000395407.2
ENST00000558684.1 |
PAQR5
|
progestin and adipoQ receptor family member V |
| chr19_+_38664224 | 0.06 |
ENST00000601054.1
|
SIPA1L3
|
signal-induced proliferation-associated 1 like 3 |
| chr14_+_77647966 | 0.06 |
ENST00000554766.1
|
TMEM63C
|
transmembrane protein 63C |
| chr20_+_17550691 | 0.06 |
ENST00000474024.1
|
DSTN
|
destrin (actin depolymerizing factor) |
| chr8_+_38758737 | 0.06 |
ENST00000521746.1
ENST00000420274.1 |
PLEKHA2
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 2 |
| chr9_+_19408919 | 0.06 |
ENST00000380376.1
|
ACER2
|
alkaline ceramidase 2 |
| chr2_-_86564740 | 0.06 |
ENST00000540790.1
ENST00000428491.1 |
REEP1
|
receptor accessory protein 1 |
| chr8_-_37594944 | 0.06 |
ENST00000330539.1
|
RP11-863K10.7
|
Uncharacterized protein |
| chr12_+_6309517 | 0.06 |
ENST00000382519.4
ENST00000009180.4 |
CD9
|
CD9 molecule |
| chr15_-_83315874 | 0.06 |
ENST00000569257.1
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr2_-_165698322 | 0.06 |
ENST00000444537.1
ENST00000414843.1 |
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr11_-_85779971 | 0.06 |
ENST00000393346.3
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr12_-_111358372 | 0.06 |
ENST00000548438.1
ENST00000228841.8 |
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow |
| chr10_+_6392278 | 0.06 |
ENST00000391437.1
|
DKFZP667F0711
|
DKFZP667F0711 |
| chr10_+_18429606 | 0.06 |
ENST00000324631.7
ENST00000352115.6 ENST00000377328.1 |
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr19_+_33865218 | 0.06 |
ENST00000585933.2
|
CEBPG
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr10_+_99258625 | 0.06 |
ENST00000370664.3
|
UBTD1
|
ubiquitin domain containing 1 |
| chr19_-_52551814 | 0.06 |
ENST00000594154.1
ENST00000598745.1 ENST00000597273.1 |
ZNF432
|
zinc finger protein 432 |
| chr2_+_173600514 | 0.06 |
ENST00000264111.6
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr4_+_89299885 | 0.06 |
ENST00000380265.5
ENST00000273960.3 |
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr5_+_44809027 | 0.06 |
ENST00000507110.1
|
MRPS30
|
mitochondrial ribosomal protein S30 |
| chr11_-_62457371 | 0.06 |
ENST00000317449.4
|
LRRN4CL
|
LRRN4 C-terminal like |
| chr1_-_6550625 | 0.06 |
ENST00000377725.1
ENST00000340850.5 |
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
| chr2_+_182850551 | 0.06 |
ENST00000452904.1
ENST00000409137.3 ENST00000280295.3 |
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr1_+_52521797 | 0.06 |
ENST00000313334.8
|
BTF3L4
|
basic transcription factor 3-like 4 |
| chr12_-_124457163 | 0.06 |
ENST00000535556.1
|
CCDC92
|
coiled-coil domain containing 92 |
| chr17_-_16256718 | 0.06 |
ENST00000476243.1
ENST00000299736.4 |
CENPV
|
centromere protein V |
| chr17_-_79869228 | 0.06 |
ENST00000570388.1
|
PCYT2
|
phosphate cytidylyltransferase 2, ethanolamine |
| chr11_+_114168085 | 0.06 |
ENST00000541754.1
|
NNMT
|
nicotinamide N-methyltransferase |
| chr11_+_46402744 | 0.06 |
ENST00000533952.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr7_-_139168402 | 0.06 |
ENST00000393039.2
|
KLRG2
|
killer cell lectin-like receptor subfamily G, member 2 |
| chr6_-_31689456 | 0.06 |
ENST00000495859.1
ENST00000375819.2 |
LY6G6C
|
lymphocyte antigen 6 complex, locus G6C |
| chr2_-_21266816 | 0.06 |
ENST00000399256.4
|
APOB
|
apolipoprotein B |
| chr17_-_4463856 | 0.06 |
ENST00000574584.1
ENST00000381550.3 ENST00000301395.3 |
GGT6
|
gamma-glutamyltransferase 6 |
| chr1_+_95582881 | 0.06 |
ENST00000370203.4
ENST00000456991.1 |
TMEM56
|
transmembrane protein 56 |
| chr12_+_57854274 | 0.06 |
ENST00000528432.1
|
GLI1
|
GLI family zinc finger 1 |
| chr11_-_3013497 | 0.06 |
ENST00000448187.1
ENST00000532325.2 ENST00000399614.2 |
NAP1L4
|
nucleosome assembly protein 1-like 4 |
| chr5_+_80256453 | 0.05 |
ENST00000265080.4
|
RASGRF2
|
Ras protein-specific guanine nucleotide-releasing factor 2 |
| chr7_-_2883928 | 0.05 |
ENST00000275364.3
|
GNA12
|
guanine nucleotide binding protein (G protein) alpha 12 |
| chr15_-_78526855 | 0.05 |
ENST00000541759.1
ENST00000558130.1 |
ACSBG1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr20_+_49348081 | 0.05 |
ENST00000371610.2
|
PARD6B
|
par-6 family cell polarity regulator beta |
| chr22_-_19974616 | 0.05 |
ENST00000344269.3
ENST00000401994.1 ENST00000406522.1 |
ARVCF
|
armadillo repeat gene deleted in velocardiofacial syndrome |
| chr16_-_30022735 | 0.05 |
ENST00000564944.1
|
DOC2A
|
double C2-like domains, alpha |
| chr6_+_54711533 | 0.05 |
ENST00000306858.7
|
FAM83B
|
family with sequence similarity 83, member B |
| chr19_-_40931891 | 0.05 |
ENST00000357949.4
|
SERTAD1
|
SERTA domain containing 1 |
| chr7_+_94139105 | 0.05 |
ENST00000297273.4
|
CASD1
|
CAS1 domain containing 1 |
| chr2_-_106054952 | 0.05 |
ENST00000336660.5
ENST00000393352.3 ENST00000607522.1 |
FHL2
|
four and a half LIM domains 2 |
| chr1_+_113051400 | 0.05 |
ENST00000369684.4
|
WNT2B
|
wingless-type MMTV integration site family, member 2B |
| chr7_+_16793160 | 0.05 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13 |
| chr18_+_21594384 | 0.05 |
ENST00000584250.1
|
TTC39C
|
tetratricopeptide repeat domain 39C |
| chr9_+_135937365 | 0.05 |
ENST00000372080.4
ENST00000351304.7 |
CEL
|
carboxyl ester lipase |
| chr14_-_104028595 | 0.05 |
ENST00000337322.4
ENST00000445922.2 |
BAG5
|
BCL2-associated athanogene 5 |
| chr10_+_135122906 | 0.05 |
ENST00000368554.4
|
ZNF511
|
zinc finger protein 511 |
| chr1_-_17445930 | 0.05 |
ENST00000375486.4
ENST00000375481.1 ENST00000444885.2 |
PADI2
|
peptidyl arginine deiminase, type II |
| chr13_+_103046954 | 0.05 |
ENST00000606448.1
|
FGF14-AS2
|
FGF14 antisense RNA 2 |
| chr19_+_10217270 | 0.05 |
ENST00000446223.1
|
PPAN
|
peter pan homolog (Drosophila) |
| chr8_-_80993010 | 0.05 |
ENST00000537855.1
ENST00000520527.1 ENST00000517427.1 ENST00000448733.2 ENST00000379097.3 |
TPD52
|
tumor protein D52 |
| chr1_-_113249678 | 0.05 |
ENST00000369633.2
ENST00000425265.2 ENST00000369632.2 ENST00000436685.2 |
RHOC
|
ras homolog family member C |
| chr19_-_18709357 | 0.05 |
ENST00000597131.1
|
CRLF1
|
cytokine receptor-like factor 1 |
| chr16_-_30032610 | 0.05 |
ENST00000574405.1
|
DOC2A
|
double C2-like domains, alpha |
| chr7_+_116165754 | 0.05 |
ENST00000405348.1
|
CAV1
|
caveolin 1, caveolae protein, 22kDa |
| chr1_-_229694406 | 0.05 |
ENST00000344517.4
|
ABCB10
|
ATP-binding cassette, sub-family B (MDR/TAP), member 10 |
| chr16_-_75529273 | 0.05 |
ENST00000390664.2
|
CHST6
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 6 |
| chr2_-_241835561 | 0.05 |
ENST00000388934.4
|
C2orf54
|
chromosome 2 open reading frame 54 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TWIST1_SNAI1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1903461 | Okazaki fragment processing involved in mitotic DNA replication(GO:1903461) |
| 0.1 | 0.2 | GO:0061193 | sensory organ boundary specification(GO:0008052) formation of organ boundary(GO:0010160) taste bud development(GO:0061193) |
| 0.0 | 0.0 | GO:0043000 | Golgi to plasma membrane CFTR protein transport(GO:0043000) |
| 0.0 | 0.1 | GO:0097195 | pilomotor reflex(GO:0097195) |
| 0.0 | 0.1 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.1 | GO:1903609 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.0 | 0.1 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.1 | GO:1902908 | regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) regulation of melanosome transport(GO:1902908) positive regulation of melanosome transport(GO:1902910) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.1 | GO:0042495 | detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.1 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.1 | GO:0051780 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.0 | 0.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.3 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.1 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.2 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.0 | 0.1 | GO:1990426 | homologous recombination-dependent replication fork processing(GO:1990426) |
| 0.0 | 0.1 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.0 | 0.1 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.1 | GO:2001226 | negative regulation of anion channel activity(GO:0010360) negative regulation of chloride transport(GO:2001226) |
| 0.0 | 0.1 | GO:1902232 | regulation of positive thymic T cell selection(GO:1902232) |
| 0.0 | 0.1 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.0 | 0.0 | GO:1903572 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.0 | 0.1 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.1 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.0 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:1902669 | positive regulation of axon guidance(GO:1902669) |
| 0.0 | 0.0 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.0 | 0.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.1 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.0 | 0.1 | GO:0034343 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.0 | 0.1 | GO:2000980 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.1 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.0 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.0 | 0.1 | GO:0036483 | neuron intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0036483) regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903381) negative regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903382) |
| 0.0 | 0.0 | GO:0060927 | Purkinje myocyte differentiation(GO:0003168) cardiac pacemaker cell fate commitment(GO:0060927) atrioventricular node cell fate commitment(GO:0060929) |
| 0.0 | 0.0 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.0 | GO:2000359 | negative regulation of fertilization(GO:0060467) regulation of binding of sperm to zona pellucida(GO:2000359) negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.0 | 0.0 | GO:0060978 | angiogenesis involved in coronary vascular morphogenesis(GO:0060978) |
| 0.0 | 0.0 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.0 | 0.1 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.0 | 0.1 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.0 | 0.1 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.0 | 0.1 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.0 | 0.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.0 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.2 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.0 | GO:0060623 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) response to DDT(GO:0046680) regulation of chromosome condensation(GO:0060623) histone H3-S10 phosphorylation involved in chromosome condensation(GO:2000775) |
| 0.0 | 0.1 | GO:0085032 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.0 | 0.0 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.0 | 0.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.0 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.0 | 0.1 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 0.0 | 0.0 | GO:0003165 | Purkinje myocyte development(GO:0003165) |
| 0.0 | 0.2 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.0 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.0 | 0.1 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.2 | GO:0030836 | positive regulation of actin filament depolymerization(GO:0030836) |
| 0.0 | 0.1 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.0 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.1 | GO:0038183 | bile acid signaling pathway(GO:0038183) |
| 0.0 | 0.1 | GO:1903433 | regulation of constitutive secretory pathway(GO:1903433) |
| 0.0 | 0.0 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.0 | GO:0060381 | regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) |
| 0.0 | 0.1 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.0 | 0.1 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.0 | 0.1 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034684 | integrin alphav-beta5 complex(GO:0034684) |
| 0.0 | 0.1 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 0.1 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.2 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.0 | GO:0097453 | mesaxon(GO:0097453) ensheathing process(GO:1990015) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.0 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.0 | 0.1 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.0 | 0.0 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.1 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 0.0 | 0.1 | GO:0000313 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.0 | 0.0 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.0 | 0.1 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0072591 | citrate-L-glutamate ligase activity(GO:0072591) |
| 0.0 | 0.3 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.2 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.2 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.1 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.0 | 0.1 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.0 | 0.1 | GO:0004961 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.0 | 0.2 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.1 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.3 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.1 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.0 | 0.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.1 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.0 | 0.1 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 0.0 | 0.1 | GO:0004306 | ethanolamine-phosphate cytidylyltransferase activity(GO:0004306) |
| 0.0 | 0.1 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.0 | 0.1 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.0 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 0.1 | GO:0030760 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.0 | 0.0 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.0 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.0 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.0 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.1 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.0 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.1 | GO:0097677 | STAT family protein binding(GO:0097677) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.3 | REACTOME PROCESSIVE SYNTHESIS ON THE LAGGING STRAND | Genes involved in Processive synthesis on the lagging strand |