Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
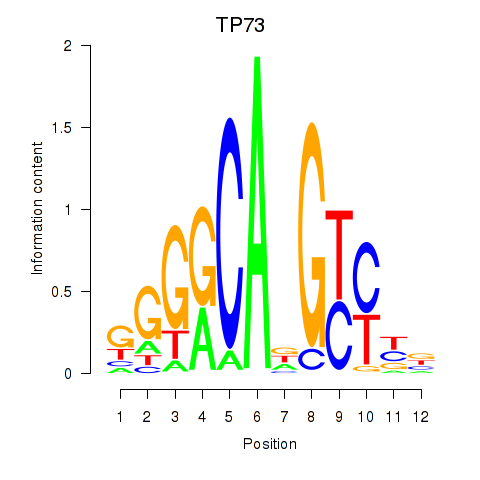
Results for TP73
Z-value: 0.20
Transcription factors associated with TP73
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TP73
|
ENSG00000078900.10 | tumor protein p73 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TP73 | hg19_v2_chr1_+_3607228_3607250 | -0.71 | 2.9e-01 | Click! |
Activity profile of TP73 motif
Sorted Z-values of TP73 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_74466744 | 0.09 |
ENST00000560862.1
ENST00000395118.1 |
ISLR
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr16_+_222846 | 0.08 |
ENST00000251595.6
ENST00000397806.1 |
HBA2
|
hemoglobin, alpha 2 |
| chrX_-_38186811 | 0.07 |
ENST00000318842.7
|
RPGR
|
retinitis pigmentosa GTPase regulator |
| chrX_-_38186775 | 0.07 |
ENST00000339363.3
ENST00000309513.3 ENST00000338898.3 ENST00000342811.3 ENST00000378505.2 |
RPGR
|
retinitis pigmentosa GTPase regulator |
| chr14_+_96858433 | 0.07 |
ENST00000267584.4
|
AK7
|
adenylate kinase 7 |
| chr20_+_35504522 | 0.06 |
ENST00000602922.1
ENST00000217320.3 |
TLDC2
|
TBC/LysM-associated domain containing 2 |
| chr12_+_113495492 | 0.05 |
ENST00000257600.3
|
DTX1
|
deltex homolog 1 (Drosophila) |
| chr16_+_84178874 | 0.05 |
ENST00000378553.5
|
DNAAF1
|
dynein, axonemal, assembly factor 1 |
| chr2_-_232328867 | 0.05 |
ENST00000453992.1
ENST00000417652.1 ENST00000454824.1 |
NCL
|
nucleolin |
| chr11_+_31833939 | 0.05 |
ENST00000530348.1
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain |
| chr9_-_128003606 | 0.05 |
ENST00000324460.6
|
HSPA5
|
heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) |
| chr3_+_100120441 | 0.05 |
ENST00000489752.1
|
LNP1
|
leukemia NUP98 fusion partner 1 |
| chr1_-_235324530 | 0.05 |
ENST00000447801.1
ENST00000366606.3 ENST00000429912.1 |
RBM34
|
RNA binding motif protein 34 |
| chr19_+_17326521 | 0.05 |
ENST00000593597.1
|
USE1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr17_+_52978185 | 0.04 |
ENST00000572405.1
ENST00000572158.1 ENST00000540336.1 ENST00000572298.1 ENST00000536554.1 ENST00000575333.1 ENST00000570499.1 ENST00000572576.1 |
TOM1L1
|
target of myb1 (chicken)-like 1 |
| chr14_-_105531759 | 0.04 |
ENST00000329797.3
ENST00000539291.2 ENST00000392585.2 |
GPR132
|
G protein-coupled receptor 132 |
| chr3_-_50340996 | 0.04 |
ENST00000266031.4
ENST00000395143.2 ENST00000457214.2 ENST00000447605.2 ENST00000418723.1 ENST00000395144.2 |
HYAL1
|
hyaluronoglucosaminidase 1 |
| chr5_+_43120985 | 0.04 |
ENST00000515326.1
|
ZNF131
|
zinc finger protein 131 |
| chr12_+_44152740 | 0.04 |
ENST00000440781.2
ENST00000431837.1 ENST00000550616.1 ENST00000448290.2 ENST00000551736.1 |
IRAK4
|
interleukin-1 receptor-associated kinase 4 |
| chr17_+_37356555 | 0.04 |
ENST00000579374.1
|
RPL19
|
ribosomal protein L19 |
| chr21_+_43442100 | 0.03 |
ENST00000455701.1
ENST00000596595.1 |
ZNF295-AS1
|
ZNF295 antisense RNA 1 |
| chr13_+_48877895 | 0.03 |
ENST00000267163.4
|
RB1
|
retinoblastoma 1 |
| chr19_+_45281118 | 0.03 |
ENST00000270279.3
ENST00000341505.4 |
CBLC
|
Cbl proto-oncogene C, E3 ubiquitin protein ligase |
| chr17_+_37356586 | 0.03 |
ENST00000579260.1
ENST00000582193.1 |
RPL19
|
ribosomal protein L19 |
| chr22_-_37976082 | 0.03 |
ENST00000215886.4
|
LGALS2
|
lectin, galactoside-binding, soluble, 2 |
| chr11_+_237016 | 0.03 |
ENST00000352303.5
|
PSMD13
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 |
| chr1_-_236228417 | 0.03 |
ENST00000264187.6
|
NID1
|
nidogen 1 |
| chr1_-_236228403 | 0.03 |
ENST00000366595.3
|
NID1
|
nidogen 1 |
| chr9_-_20382446 | 0.03 |
ENST00000380321.1
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr13_-_48877795 | 0.03 |
ENST00000436963.1
ENST00000433480.2 |
LINC00441
|
long intergenic non-protein coding RNA 441 |
| chr11_-_17035943 | 0.03 |
ENST00000355661.3
ENST00000532079.1 ENST00000448080.2 ENST00000531066.1 |
PLEKHA7
|
pleckstrin homology domain containing, family A member 7 |
| chr14_+_105190514 | 0.03 |
ENST00000330877.2
|
ADSSL1
|
adenylosuccinate synthase like 1 |
| chr3_-_100120223 | 0.03 |
ENST00000284320.5
|
TOMM70A
|
translocase of outer mitochondrial membrane 70 homolog A (S. cerevisiae) |
| chr16_+_21312170 | 0.03 |
ENST00000338573.5
ENST00000561968.1 |
CRYM-AS1
|
CRYM antisense RNA 1 |
| chr11_+_65687158 | 0.03 |
ENST00000532933.1
|
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr11_+_236959 | 0.03 |
ENST00000431206.2
ENST00000528906.1 |
PSMD13
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 |
| chr1_-_220219775 | 0.03 |
ENST00000609181.1
|
EPRS
|
glutamyl-prolyl-tRNA synthetase |
| chr3_+_136676707 | 0.02 |
ENST00000329582.4
|
IL20RB
|
interleukin 20 receptor beta |
| chr10_-_47173994 | 0.02 |
ENST00000414655.2
ENST00000545298.1 ENST00000359178.4 ENST00000358140.4 ENST00000503031.1 |
ANXA8L1
LINC00842
|
annexin A8-like 1 long intergenic non-protein coding RNA 842 |
| chr9_+_116343192 | 0.02 |
ENST00000471324.2
|
RGS3
|
regulator of G-protein signaling 3 |
| chr16_-_14724057 | 0.02 |
ENST00000539279.1
ENST00000420015.2 ENST00000437198.2 |
PARN
|
poly(A)-specific ribonuclease |
| chr10_-_97200772 | 0.02 |
ENST00000371241.1
ENST00000354106.3 ENST00000371239.1 ENST00000361941.3 ENST00000277982.5 ENST00000371245.3 |
SORBS1
|
sorbin and SH3 domain containing 1 |
| chrX_+_9880590 | 0.02 |
ENST00000452575.1
|
SHROOM2
|
shroom family member 2 |
| chr6_+_155537771 | 0.02 |
ENST00000275246.7
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr6_-_33256664 | 0.02 |
ENST00000444176.1
|
WDR46
|
WD repeat domain 46 |
| chrX_+_77166172 | 0.02 |
ENST00000343533.5
ENST00000350425.4 ENST00000341514.6 |
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr7_-_100493744 | 0.02 |
ENST00000428317.1
ENST00000441605.1 |
ACHE
|
acetylcholinesterase (Yt blood group) |
| chr15_+_41136369 | 0.02 |
ENST00000563656.1
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr12_+_102271436 | 0.02 |
ENST00000544152.1
|
DRAM1
|
DNA-damage regulated autophagy modulator 1 |
| chr5_+_119799927 | 0.02 |
ENST00000407149.2
ENST00000379551.2 |
PRR16
|
proline rich 16 |
| chr10_-_91295304 | 0.02 |
ENST00000341233.4
ENST00000371790.4 |
SLC16A12
|
solute carrier family 16, member 12 |
| chr9_+_6758024 | 0.02 |
ENST00000442236.2
|
KDM4C
|
lysine (K)-specific demethylase 4C |
| chr20_-_49253425 | 0.02 |
ENST00000045083.2
|
FAM65C
|
family with sequence similarity 65, member C |
| chr17_-_39459103 | 0.02 |
ENST00000391353.1
|
KRTAP29-1
|
keratin associated protein 29-1 |
| chr11_+_236540 | 0.02 |
ENST00000532097.1
|
PSMD13
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 |
| chr7_-_100493482 | 0.02 |
ENST00000411582.1
ENST00000419336.2 ENST00000241069.5 ENST00000302913.4 |
ACHE
|
acetylcholinesterase (Yt blood group) |
| chr2_+_95940186 | 0.02 |
ENST00000403131.2
ENST00000317668.4 ENST00000317620.9 |
PROM2
|
prominin 2 |
| chr17_-_47492164 | 0.02 |
ENST00000512041.2
ENST00000446735.1 ENST00000504124.1 |
PHB
|
prohibitin |
| chr2_+_220283091 | 0.02 |
ENST00000373960.3
|
DES
|
desmin |
| chr15_+_41136586 | 0.02 |
ENST00000431806.1
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr14_-_25519095 | 0.02 |
ENST00000419632.2
ENST00000358326.2 ENST00000396700.1 ENST00000548724.1 |
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chr8_+_120885949 | 0.02 |
ENST00000523492.1
ENST00000286234.5 |
DEPTOR
|
DEP domain containing MTOR-interacting protein |
| chr12_-_82752565 | 0.02 |
ENST00000256151.7
|
CCDC59
|
coiled-coil domain containing 59 |
| chr6_-_112194484 | 0.02 |
ENST00000518295.1
ENST00000484067.2 ENST00000229470.5 ENST00000356013.2 ENST00000368678.4 ENST00000523238.1 ENST00000354650.3 |
FYN
|
FYN oncogene related to SRC, FGR, YES |
| chr12_-_82752159 | 0.02 |
ENST00000552377.1
|
CCDC59
|
coiled-coil domain containing 59 |
| chr11_-_31833719 | 0.01 |
ENST00000524853.1
|
PAX6
|
paired box 6 |
| chrX_+_153770421 | 0.01 |
ENST00000369609.5
ENST00000369607.1 |
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr11_+_75479850 | 0.01 |
ENST00000376262.3
ENST00000604733.1 |
DGAT2
|
diacylglycerol O-acyltransferase 2 |
| chr15_+_41136216 | 0.01 |
ENST00000562057.1
ENST00000344051.4 |
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr17_-_47492236 | 0.01 |
ENST00000434917.2
ENST00000300408.3 ENST00000511832.1 ENST00000419140.2 |
PHB
|
prohibitin |
| chr22_+_29168652 | 0.01 |
ENST00000249064.4
ENST00000444523.1 ENST00000448492.2 ENST00000421503.2 |
CCDC117
|
coiled-coil domain containing 117 |
| chr10_-_99185798 | 0.01 |
ENST00000439965.2
|
AL355490.1
|
LOC644215 protein; Uncharacterized protein |
| chr14_+_105935396 | 0.01 |
ENST00000494981.1
|
MTA1
|
metastasis associated 1 |
| chr11_+_65686952 | 0.01 |
ENST00000527119.1
|
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr20_-_52790055 | 0.01 |
ENST00000395955.3
|
CYP24A1
|
cytochrome P450, family 24, subfamily A, polypeptide 1 |
| chr4_-_76598544 | 0.01 |
ENST00000515457.1
ENST00000357854.3 |
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr1_-_52344416 | 0.01 |
ENST00000544028.1
|
NRD1
|
nardilysin (N-arginine dibasic convertase) |
| chr14_+_103566481 | 0.01 |
ENST00000380069.3
|
EXOC3L4
|
exocyst complex component 3-like 4 |
| chr9_-_138391692 | 0.01 |
ENST00000429260.2
|
C9orf116
|
chromosome 9 open reading frame 116 |
| chr9_+_6757634 | 0.01 |
ENST00000543771.1
ENST00000401787.3 ENST00000381306.3 ENST00000381309.3 |
KDM4C
|
lysine (K)-specific demethylase 4C |
| chr12_-_56881909 | 0.01 |
ENST00000461077.1
|
GLS2
|
glutaminase 2 (liver, mitochondrial) |
| chr6_+_35995552 | 0.01 |
ENST00000468133.1
|
MAPK14
|
mitogen-activated protein kinase 14 |
| chr12_+_82752283 | 0.01 |
ENST00000548200.1
|
METTL25
|
methyltransferase like 25 |
| chr1_+_156785425 | 0.01 |
ENST00000392302.2
|
NTRK1
|
neurotrophic tyrosine kinase, receptor, type 1 |
| chr16_-_8891481 | 0.01 |
ENST00000333050.6
|
TMEM186
|
transmembrane protein 186 |
| chr1_-_241799232 | 0.01 |
ENST00000366553.1
|
CHML
|
choroideremia-like (Rab escort protein 2) |
| chr1_-_160832642 | 0.01 |
ENST00000368034.4
|
CD244
|
CD244 molecule, natural killer cell receptor 2B4 |
| chr19_+_19627026 | 0.01 |
ENST00000608404.1
ENST00000555938.1 ENST00000503283.1 ENST00000512771.3 ENST00000428459.2 |
YJEFN3
CTC-260F20.3
NDUFA13
|
YjeF N-terminal domain containing 3 Uncharacterized protein NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 13 |
| chr1_+_110082487 | 0.01 |
ENST00000527748.1
|
GPR61
|
G protein-coupled receptor 61 |
| chrX_+_9880412 | 0.01 |
ENST00000418909.2
|
SHROOM2
|
shroom family member 2 |
| chr7_+_116502527 | 0.01 |
ENST00000361183.3
|
CAPZA2
|
capping protein (actin filament) muscle Z-line, alpha 2 |
| chr10_+_70661014 | 0.01 |
ENST00000373585.3
|
DDX50
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 50 |
| chr18_-_28681950 | 0.01 |
ENST00000251081.6
|
DSC2
|
desmocollin 2 |
| chr1_-_52344471 | 0.01 |
ENST00000352171.7
ENST00000354831.7 |
NRD1
|
nardilysin (N-arginine dibasic convertase) |
| chr1_-_220220000 | 0.01 |
ENST00000366923.3
|
EPRS
|
glutamyl-prolyl-tRNA synthetase |
| chr9_+_131445928 | 0.01 |
ENST00000372692.4
|
SET
|
SET nuclear oncogene |
| chr2_+_128293323 | 0.01 |
ENST00000389524.4
ENST00000428314.1 |
MYO7B
|
myosin VIIB |
| chr15_-_74043816 | 0.01 |
ENST00000379822.4
|
C15orf59
|
chromosome 15 open reading frame 59 |
| chr9_+_6758109 | 0.01 |
ENST00000536108.1
|
KDM4C
|
lysine (K)-specific demethylase 4C |
| chr2_-_230135937 | 0.01 |
ENST00000392054.3
ENST00000409462.1 ENST00000392055.3 |
PID1
|
phosphotyrosine interaction domain containing 1 |
| chr10_-_105238997 | 0.01 |
ENST00000369783.4
|
CALHM3
|
calcium homeostasis modulator 3 |
| chr12_+_31079362 | 0.01 |
ENST00000545802.1
|
TSPAN11
|
tetraspanin 11 |
| chr20_-_52790512 | 0.01 |
ENST00000216862.3
|
CYP24A1
|
cytochrome P450, family 24, subfamily A, polypeptide 1 |
| chr12_-_56881874 | 0.01 |
ENST00000539272.1
|
GLS2
|
glutaminase 2 (liver, mitochondrial) |
| chr17_-_79818354 | 0.01 |
ENST00000576541.1
ENST00000576380.1 ENST00000571617.1 ENST00000576052.1 ENST00000576390.1 ENST00000573778.2 ENST00000439918.2 ENST00000574914.1 ENST00000331483.4 |
P4HB
|
prolyl 4-hydroxylase, beta polypeptide |
| chr12_+_82752647 | 0.01 |
ENST00000550058.1
|
METTL25
|
methyltransferase like 25 |
| chr8_+_54764346 | 0.01 |
ENST00000297313.3
ENST00000344277.6 |
RGS20
|
regulator of G-protein signaling 20 |
| chr3_+_136676851 | 0.01 |
ENST00000309741.5
|
IL20RB
|
interleukin 20 receptor beta |
| chr17_+_37356528 | 0.01 |
ENST00000225430.4
|
RPL19
|
ribosomal protein L19 |
| chr5_+_78532003 | 0.01 |
ENST00000396137.4
|
JMY
|
junction mediating and regulatory protein, p53 cofactor |
| chr7_-_2883650 | 0.01 |
ENST00000544127.1
|
GNA12
|
guanine nucleotide binding protein (G protein) alpha 12 |
| chr11_+_63448918 | 0.00 |
ENST00000341307.2
ENST00000356000.3 ENST00000542238.1 |
RTN3
|
reticulon 3 |
| chr19_+_44037546 | 0.00 |
ENST00000601282.1
|
ZNF575
|
zinc finger protein 575 |
| chr12_+_82752275 | 0.00 |
ENST00000248306.3
|
METTL25
|
methyltransferase like 25 |
| chr7_-_128045984 | 0.00 |
ENST00000470772.1
ENST00000480861.1 ENST00000496200.1 |
IMPDH1
|
IMP (inosine 5'-monophosphate) dehydrogenase 1 |
| chr6_-_122792919 | 0.00 |
ENST00000339697.4
|
SERINC1
|
serine incorporator 1 |
| chr16_+_8891670 | 0.00 |
ENST00000268261.4
ENST00000539622.1 ENST00000569958.1 ENST00000537352.1 |
PMM2
|
phosphomannomutase 2 |
| chr3_+_48481658 | 0.00 |
ENST00000438607.2
|
TMA7
|
translation machinery associated 7 homolog (S. cerevisiae) |
| chr16_+_5008290 | 0.00 |
ENST00000251170.7
|
SEC14L5
|
SEC14-like 5 (S. cerevisiae) |
| chr14_-_25519317 | 0.00 |
ENST00000323944.5
|
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chr16_-_84178728 | 0.00 |
ENST00000562224.1
ENST00000434463.3 ENST00000564998.1 ENST00000219439.4 |
HSDL1
|
hydroxysteroid dehydrogenase like 1 |
| chr1_-_111148241 | 0.00 |
ENST00000440270.1
|
KCNA2
|
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr3_+_42201653 | 0.00 |
ENST00000341421.3
ENST00000396175.1 |
TRAK1
|
trafficking protein, kinesin binding 1 |
| chr1_+_162467595 | 0.00 |
ENST00000538489.1
ENST00000489294.1 |
UHMK1
|
U2AF homology motif (UHM) kinase 1 |
| chr7_-_2883928 | 0.00 |
ENST00000275364.3
|
GNA12
|
guanine nucleotide binding protein (G protein) alpha 12 |
| chr8_-_70745575 | 0.00 |
ENST00000524945.1
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1 |
| chr2_-_158485387 | 0.00 |
ENST00000243349.8
|
ACVR1C
|
activin A receptor, type IC |
| chr11_+_63448955 | 0.00 |
ENST00000377819.5
ENST00000339997.4 ENST00000540798.1 ENST00000545432.1 ENST00000543552.1 ENST00000537981.1 |
RTN3
|
reticulon 3 |
| chr1_-_78444776 | 0.00 |
ENST00000370767.1
ENST00000421641.1 |
FUBP1
|
far upstream element (FUSE) binding protein 1 |
| chr17_-_42452063 | 0.00 |
ENST00000588098.1
|
ITGA2B
|
integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41) |
| chr15_+_64680003 | 0.00 |
ENST00000261884.3
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr1_+_204494618 | 0.00 |
ENST00000367180.1
ENST00000391947.2 |
MDM4
|
Mdm4 p53 binding protein homolog (mouse) |
| chrX_-_138724994 | 0.00 |
ENST00000536274.1
|
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr14_+_96858454 | 0.00 |
ENST00000555570.1
|
AK7
|
adenylate kinase 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TP73
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.0 | 0.0 | GO:0045212 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 | 0.0 | GO:0006433 | glutamyl-tRNA aminoacylation(GO:0006424) prolyl-tRNA aminoacylation(GO:0006433) |
| 0.0 | 0.0 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.0 | GO:0008057 | eye pigment granule organization(GO:0008057) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.0 | 0.0 | GO:0035189 | Rb-E2F complex(GO:0035189) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.0 | GO:0004827 | glutamate-tRNA ligase activity(GO:0004818) proline-tRNA ligase activity(GO:0004827) |
| 0.0 | 0.0 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |