Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
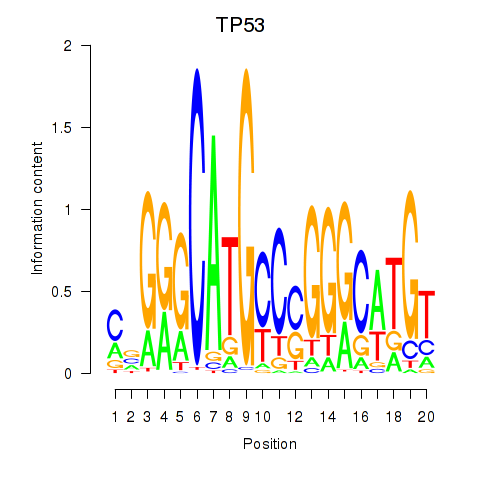
Results for TP53
Z-value: 0.85
Transcription factors associated with TP53
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TP53
|
ENSG00000141510.11 | tumor protein p53 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TP53 | hg19_v2_chr17_-_7590745_7590856 | 0.91 | 8.6e-02 | Click! |
Activity profile of TP53 motif
Sorted Z-values of TP53 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_47551836 | 0.59 |
ENST00000253047.6
|
TMEM160
|
transmembrane protein 160 |
| chr19_-_47735918 | 0.56 |
ENST00000449228.1
ENST00000300880.7 ENST00000341983.4 |
BBC3
|
BCL2 binding component 3 |
| chr8_+_22429205 | 0.49 |
ENST00000520207.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr2_+_233734994 | 0.46 |
ENST00000331342.2
|
C2orf82
|
chromosome 2 open reading frame 82 |
| chr17_+_73455788 | 0.46 |
ENST00000581519.1
|
KIAA0195
|
KIAA0195 |
| chr17_+_76210367 | 0.44 |
ENST00000592734.1
ENST00000587746.1 |
BIRC5
|
baculoviral IAP repeat containing 5 |
| chr11_+_64126671 | 0.43 |
ENST00000530504.1
|
RPS6KA4
|
ribosomal protein S6 kinase, 90kDa, polypeptide 4 |
| chr1_-_201438282 | 0.41 |
ENST00000367311.3
ENST00000367309.1 |
PHLDA3
|
pleckstrin homology-like domain, family A, member 3 |
| chr6_+_33378517 | 0.40 |
ENST00000428274.1
|
PHF1
|
PHD finger protein 1 |
| chr1_+_11796177 | 0.40 |
ENST00000400895.2
ENST00000376629.4 ENST00000376627.2 ENST00000314340.5 ENST00000452018.2 ENST00000510878.1 |
AGTRAP
|
angiotensin II receptor-associated protein |
| chr1_+_11796126 | 0.40 |
ENST00000376637.3
|
AGTRAP
|
angiotensin II receptor-associated protein |
| chr16_-_29874211 | 0.37 |
ENST00000563415.1
|
CDIPT
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase |
| chr11_+_64879317 | 0.37 |
ENST00000526809.1
ENST00000279263.7 ENST00000524986.1 ENST00000534371.1 ENST00000540748.1 ENST00000525385.1 ENST00000345348.5 ENST00000531321.1 ENST00000529414.1 ENST00000526085.1 ENST00000530750.1 |
TM7SF2
|
transmembrane 7 superfamily member 2 |
| chr17_+_14204389 | 0.33 |
ENST00000360954.2
|
HS3ST3B1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1 |
| chr20_+_4129496 | 0.32 |
ENST00000346595.2
|
SMOX
|
spermine oxidase |
| chr11_+_64126614 | 0.32 |
ENST00000528057.1
ENST00000334205.4 ENST00000294261.4 |
RPS6KA4
|
ribosomal protein S6 kinase, 90kDa, polypeptide 4 |
| chr22_+_21336267 | 0.31 |
ENST00000215739.8
|
LZTR1
|
leucine-zipper-like transcription regulator 1 |
| chr11_+_64073699 | 0.31 |
ENST00000405666.1
ENST00000468670.1 |
ESRRA
|
estrogen-related receptor alpha |
| chr1_+_151584544 | 0.29 |
ENST00000458013.2
ENST00000368843.3 |
SNX27
|
sorting nexin family member 27 |
| chr11_-_44972476 | 0.29 |
ENST00000527685.1
ENST00000308212.5 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr16_+_2570340 | 0.28 |
ENST00000568263.1
ENST00000293971.6 ENST00000302956.4 ENST00000413459.3 ENST00000566706.1 ENST00000569879.1 |
AMDHD2
|
amidohydrolase domain containing 2 |
| chr7_+_140396946 | 0.28 |
ENST00000476470.1
ENST00000471136.1 |
NDUFB2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa |
| chr7_+_140396756 | 0.28 |
ENST00000460088.1
ENST00000472695.1 |
NDUFB2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa |
| chr19_+_13228917 | 0.27 |
ENST00000586171.1
|
NACC1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr17_-_61523622 | 0.27 |
ENST00000448884.2
ENST00000582297.1 ENST00000582034.1 ENST00000578072.1 ENST00000360793.3 |
CYB561
|
cytochrome b561 |
| chr22_-_37415475 | 0.27 |
ENST00000403892.3
ENST00000249042.3 ENST00000438203.1 |
TST
|
thiosulfate sulfurtransferase (rhodanese) |
| chr22_+_40766582 | 0.27 |
ENST00000457767.1
ENST00000248929.9 ENST00000454798.2 |
SGSM3
|
small G protein signaling modulator 3 |
| chr20_-_23618582 | 0.27 |
ENST00000398411.1
ENST00000376925.3 |
CST3
|
cystatin C |
| chr1_-_54518865 | 0.27 |
ENST00000371337.3
|
TMEM59
|
transmembrane protein 59 |
| chr22_-_20307532 | 0.26 |
ENST00000405465.3
ENST00000248879.3 |
DGCR6L
|
DiGeorge syndrome critical region gene 6-like |
| chr17_+_73089382 | 0.26 |
ENST00000538213.2
ENST00000584118.1 |
SLC16A5
|
solute carrier family 16 (monocarboxylate transporter), member 5 |
| chr19_-_42721819 | 0.25 |
ENST00000336034.4
ENST00000598200.1 ENST00000598727.1 ENST00000596251.1 |
DEDD2
|
death effector domain containing 2 |
| chr7_+_140396465 | 0.25 |
ENST00000476279.1
ENST00000247866.4 ENST00000461457.1 ENST00000465506.1 ENST00000204307.5 ENST00000464566.1 |
NDUFB2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa |
| chr12_-_113772835 | 0.25 |
ENST00000552014.1
ENST00000548186.1 ENST00000202831.3 ENST00000549181.1 |
SLC8B1
|
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
| chr16_-_31106211 | 0.25 |
ENST00000532364.1
ENST00000529564.1 ENST00000319788.7 ENST00000354895.4 ENST00000394975.2 |
RP11-196G11.1
VKORC1
|
Uncharacterized protein vitamin K epoxide reductase complex, subunit 1 |
| chr16_-_29874462 | 0.24 |
ENST00000566113.1
ENST00000569956.1 ENST00000570016.1 |
CDIPT
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase |
| chr15_-_72767490 | 0.24 |
ENST00000565181.1
|
RP11-1007O24.3
|
RP11-1007O24.3 |
| chr19_+_54496132 | 0.23 |
ENST00000346968.2
|
CACNG6
|
calcium channel, voltage-dependent, gamma subunit 6 |
| chr19_+_56652643 | 0.23 |
ENST00000586123.1
|
ZNF444
|
zinc finger protein 444 |
| chr11_-_44972418 | 0.23 |
ENST00000525680.1
ENST00000528290.1 ENST00000530035.1 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr12_+_53895052 | 0.22 |
ENST00000552857.1
|
TARBP2
|
TAR (HIV-1) RNA binding protein 2 |
| chr11_+_119039414 | 0.22 |
ENST00000409991.1
ENST00000292199.2 ENST00000409265.4 ENST00000409109.1 |
NLRX1
|
NLR family member X1 |
| chr22_+_42949925 | 0.22 |
ENST00000327678.5
ENST00000340239.4 ENST00000407614.4 ENST00000335879.5 |
SERHL2
|
serine hydrolase-like 2 |
| chr19_-_6591113 | 0.22 |
ENST00000423145.3
ENST00000245903.3 |
CD70
|
CD70 molecule |
| chr19_+_56653064 | 0.21 |
ENST00000593100.1
|
ZNF444
|
zinc finger protein 444 |
| chr15_+_89164560 | 0.21 |
ENST00000379231.3
ENST00000559528.1 |
AEN
|
apoptosis enhancing nuclease |
| chr22_-_28392227 | 0.21 |
ENST00000431039.1
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr10_-_105212059 | 0.21 |
ENST00000260743.5
|
CALHM2
|
calcium homeostasis modulator 2 |
| chr16_-_68000717 | 0.21 |
ENST00000541864.2
|
SLC12A4
|
solute carrier family 12 (potassium/chloride transporter), member 4 |
| chr17_-_45918539 | 0.21 |
ENST00000584123.1
ENST00000578323.1 ENST00000407215.3 ENST00000290216.9 |
SCRN2
|
secernin 2 |
| chr12_+_56546363 | 0.21 |
ENST00000551834.1
ENST00000552568.1 |
MYL6B
|
myosin, light chain 6B, alkali, smooth muscle and non-muscle |
| chr17_+_18128896 | 0.21 |
ENST00000316843.4
|
LLGL1
|
lethal giant larvae homolog 1 (Drosophila) |
| chr11_-_44972390 | 0.21 |
ENST00000395648.3
ENST00000531928.2 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr7_-_75241096 | 0.20 |
ENST00000420909.1
|
HIP1
|
huntingtin interacting protein 1 |
| chr6_+_33378738 | 0.20 |
ENST00000374512.3
ENST00000374516.3 |
PHF1
|
PHD finger protein 1 |
| chr16_+_57653989 | 0.20 |
ENST00000567835.1
ENST00000569372.1 ENST00000563548.1 ENST00000562003.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr11_+_289155 | 0.20 |
ENST00000409655.1
|
ATHL1
|
ATH1, acid trehalase-like 1 (yeast) |
| chr19_-_5567842 | 0.20 |
ENST00000587632.1
|
TINCR
|
tissue differentiation-inducing non-protein coding RNA |
| chr16_-_31106048 | 0.19 |
ENST00000300851.6
|
VKORC1
|
vitamin K epoxide reductase complex, subunit 1 |
| chr11_-_44972299 | 0.19 |
ENST00000528473.1
|
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr10_-_73611046 | 0.19 |
ENST00000394934.1
ENST00000394936.3 |
PSAP
|
prosaposin |
| chr8_+_95731904 | 0.19 |
ENST00000522422.1
|
DPY19L4
|
dpy-19-like 4 (C. elegans) |
| chr1_-_209825674 | 0.18 |
ENST00000367030.3
ENST00000356082.4 |
LAMB3
|
laminin, beta 3 |
| chr19_+_489140 | 0.18 |
ENST00000587541.1
|
MADCAM1
|
mucosal vascular addressin cell adhesion molecule 1 |
| chr11_-_73694346 | 0.18 |
ENST00000310473.3
|
UCP2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr19_-_1605424 | 0.18 |
ENST00000589880.1
ENST00000585671.1 ENST00000591899.3 |
UQCR11
|
ubiquinol-cytochrome c reductase, complex III subunit XI |
| chr10_-_105212141 | 0.18 |
ENST00000369788.3
|
CALHM2
|
calcium homeostasis modulator 2 |
| chrX_+_192989 | 0.17 |
ENST00000399012.1
ENST00000430923.2 |
PLCXD1
|
phosphatidylinositol-specific phospholipase C, X domain containing 1 |
| chr2_+_10281509 | 0.17 |
ENST00000381786.3
|
C2orf48
|
chromosome 2 open reading frame 48 |
| chr16_+_57653854 | 0.17 |
ENST00000568908.1
ENST00000568909.1 ENST00000566778.1 ENST00000561988.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr1_-_156051789 | 0.17 |
ENST00000532414.2
|
MEX3A
|
mex-3 RNA binding family member A |
| chr17_-_61523535 | 0.17 |
ENST00000584031.1
ENST00000392976.1 |
CYB561
|
cytochrome b561 |
| chr19_+_56652686 | 0.17 |
ENST00000592949.1
|
ZNF444
|
zinc finger protein 444 |
| chr19_+_49458107 | 0.17 |
ENST00000539787.1
ENST00000345358.7 ENST00000391871.3 ENST00000415969.2 ENST00000354470.3 ENST00000506183.1 ENST00000293288.8 |
BAX
|
BCL2-associated X protein |
| chr19_+_13988061 | 0.17 |
ENST00000339133.5
ENST00000397555.2 |
NANOS3
|
nanos homolog 3 (Drosophila) |
| chr20_+_62289640 | 0.17 |
ENST00000508582.2
ENST00000360203.5 ENST00000356810.4 |
RTEL1
|
regulator of telomere elongation helicase 1 |
| chr1_-_1243392 | 0.17 |
ENST00000354700.5
|
ACAP3
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3 |
| chr19_-_45735138 | 0.17 |
ENST00000252482.3
|
EXOC3L2
|
exocyst complex component 3-like 2 |
| chr19_+_56652556 | 0.16 |
ENST00000337080.3
|
ZNF444
|
zinc finger protein 444 |
| chr11_+_62538775 | 0.16 |
ENST00000294168.3
ENST00000526261.1 |
TAF6L
|
TAF6-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor, 65kDa |
| chr9_-_135996537 | 0.16 |
ENST00000372050.3
ENST00000372047.3 |
RALGDS
|
ral guanine nucleotide dissociation stimulator |
| chr11_+_289110 | 0.16 |
ENST00000409548.2
|
ATHL1
|
ATH1, acid trehalase-like 1 (yeast) |
| chr11_-_65626797 | 0.16 |
ENST00000525451.2
|
CFL1
|
cofilin 1 (non-muscle) |
| chr20_+_35234137 | 0.16 |
ENST00000344795.3
ENST00000373852.5 |
C20orf24
|
chromosome 20 open reading frame 24 |
| chr22_-_18257178 | 0.16 |
ENST00000342111.5
|
BID
|
BH3 interacting domain death agonist |
| chr12_+_53895364 | 0.16 |
ENST00000552817.1
ENST00000394357.2 |
TARBP2
|
TAR (HIV-1) RNA binding protein 2 |
| chr1_-_202936394 | 0.16 |
ENST00000367249.4
|
CYB5R1
|
cytochrome b5 reductase 1 |
| chr15_-_30685563 | 0.16 |
ENST00000401522.3
|
CHRFAM7A
|
CHRNA7 (cholinergic receptor, nicotinic, alpha 7, exons 5-10) and FAM7A (family with sequence similarity 7A, exons A-E) fusion |
| chr10_+_134000404 | 0.15 |
ENST00000338492.4
ENST00000368629.1 |
DPYSL4
|
dihydropyrimidinase-like 4 |
| chr20_+_33759854 | 0.15 |
ENST00000216968.4
|
PROCR
|
protein C receptor, endothelial |
| chr1_+_27153173 | 0.15 |
ENST00000374142.4
|
ZDHHC18
|
zinc finger, DHHC-type containing 18 |
| chr16_-_3422283 | 0.15 |
ENST00000399974.3
|
MTRNR2L4
|
MT-RNR2-like 4 |
| chr12_+_53645870 | 0.15 |
ENST00000329548.4
|
MFSD5
|
major facilitator superfamily domain containing 5 |
| chr20_-_1373606 | 0.15 |
ENST00000381715.1
ENST00000439640.2 ENST00000381719.3 |
FKBP1A
|
FK506 binding protein 1A, 12kDa |
| chr16_+_2961973 | 0.15 |
ENST00000399667.2
ENST00000416288.2 ENST00000570425.1 ENST00000573525.1 |
FLYWCH1
|
FLYWCH-type zinc finger 1 |
| chr11_+_827553 | 0.15 |
ENST00000528542.2
ENST00000450448.1 |
EFCAB4A
|
EF-hand calcium binding domain 4A |
| chr14_-_64971288 | 0.15 |
ENST00000394715.1
|
ZBTB25
|
zinc finger and BTB domain containing 25 |
| chr11_-_47546250 | 0.14 |
ENST00000543178.1
|
CELF1
|
CUGBP, Elav-like family member 1 |
| chr16_-_67997947 | 0.14 |
ENST00000537830.2
|
SLC12A4
|
solute carrier family 12 (potassium/chloride transporter), member 4 |
| chr20_+_4129426 | 0.14 |
ENST00000339123.6
ENST00000305958.4 ENST00000278795.3 |
SMOX
|
spermine oxidase |
| chr14_-_20929624 | 0.14 |
ENST00000398020.4
ENST00000250489.4 |
TMEM55B
|
transmembrane protein 55B |
| chr2_+_47168630 | 0.14 |
ENST00000263737.6
|
TTC7A
|
tetratricopeptide repeat domain 7A |
| chr22_+_38302285 | 0.14 |
ENST00000215957.6
|
MICALL1
|
MICAL-like 1 |
| chr9_-_140115775 | 0.14 |
ENST00000391553.1
ENST00000392827.1 |
RNF208
|
ring finger protein 208 |
| chr1_-_1243252 | 0.14 |
ENST00000353662.3
|
ACAP3
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3 |
| chr17_+_76210267 | 0.14 |
ENST00000301633.4
ENST00000350051.3 ENST00000374948.2 ENST00000590449.1 |
BIRC5
|
baculoviral IAP repeat containing 5 |
| chr19_+_13229126 | 0.14 |
ENST00000292431.4
|
NACC1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr8_-_22926623 | 0.14 |
ENST00000276431.4
|
TNFRSF10B
|
tumor necrosis factor receptor superfamily, member 10b |
| chr8_-_12293852 | 0.14 |
ENST00000262365.4
ENST00000351291.4 ENST00000309608.5 ENST00000527331.1 ENST00000532480.1 ENST00000393715.3 |
FAM86B2
|
family with sequence similarity 86, member B2 |
| chr4_+_103422499 | 0.13 |
ENST00000511926.1
ENST00000507079.1 |
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| chrX_-_153285251 | 0.13 |
ENST00000444230.1
ENST00000393682.1 ENST00000393687.2 ENST00000429936.2 ENST00000369974.2 |
IRAK1
|
interleukin-1 receptor-associated kinase 1 |
| chr17_-_14140166 | 0.13 |
ENST00000420162.2
ENST00000431716.2 |
CDRT15
|
CMT1A duplicated region transcript 15 |
| chr17_-_7387524 | 0.13 |
ENST00000311403.4
|
ZBTB4
|
zinc finger and BTB domain containing 4 |
| chr14_-_64970494 | 0.13 |
ENST00000608382.1
|
ZBTB25
|
zinc finger and BTB domain containing 25 |
| chrX_-_153285395 | 0.13 |
ENST00000369980.3
|
IRAK1
|
interleukin-1 receptor-associated kinase 1 |
| chr12_+_53894672 | 0.12 |
ENST00000266987.2
ENST00000456234.2 |
TARBP2
|
TAR (HIV-1) RNA binding protein 2 |
| chr1_+_116519223 | 0.12 |
ENST00000369502.1
|
SLC22A15
|
solute carrier family 22, member 15 |
| chr7_-_72439997 | 0.12 |
ENST00000285805.3
|
TRIM74
|
tripartite motif containing 74 |
| chr9_+_139846708 | 0.12 |
ENST00000371633.3
|
LCN12
|
lipocalin 12 |
| chr22_-_39239987 | 0.12 |
ENST00000333039.2
|
NPTXR
|
neuronal pentraxin receptor |
| chr9_+_35605274 | 0.12 |
ENST00000336395.5
|
TESK1
|
testis-specific kinase 1 |
| chr12_-_6451235 | 0.12 |
ENST00000440083.2
ENST00000162749.2 |
TNFRSF1A
|
tumor necrosis factor receptor superfamily, member 1A |
| chr14_-_75078725 | 0.12 |
ENST00000556690.1
|
LTBP2
|
latent transforming growth factor beta binding protein 2 |
| chr11_-_64570706 | 0.12 |
ENST00000294066.2
ENST00000377350.3 |
MAP4K2
|
mitogen-activated protein kinase kinase kinase kinase 2 |
| chr1_+_2477831 | 0.12 |
ENST00000606645.1
|
RP3-395M20.12
|
RP3-395M20.12 |
| chr11_+_60699222 | 0.12 |
ENST00000536409.1
|
TMEM132A
|
transmembrane protein 132A |
| chrX_-_54209640 | 0.12 |
ENST00000375180.2
ENST00000328235.4 ENST00000477084.1 |
FAM120C
|
family with sequence similarity 120C |
| chr6_+_157802165 | 0.11 |
ENST00000414563.2
ENST00000359775.5 |
ZDHHC14
|
zinc finger, DHHC-type containing 14 |
| chr19_+_42746927 | 0.11 |
ENST00000378108.1
|
AC006486.1
|
AC006486.1 |
| chr7_-_100287071 | 0.11 |
ENST00000275732.5
|
GIGYF1
|
GRB10 interacting GYF protein 1 |
| chr7_-_56101826 | 0.11 |
ENST00000421626.1
|
PSPH
|
phosphoserine phosphatase |
| chr11_+_706595 | 0.11 |
ENST00000531348.1
ENST00000530636.1 |
EPS8L2
|
EPS8-like 2 |
| chr12_-_6451186 | 0.11 |
ENST00000540022.1
ENST00000536194.1 |
TNFRSF1A
|
tumor necrosis factor receptor superfamily, member 1A |
| chr10_+_11784360 | 0.11 |
ENST00000379215.4
ENST00000420401.1 |
ECHDC3
|
enoyl CoA hydratase domain containing 3 |
| chr22_+_29279552 | 0.11 |
ENST00000544604.2
|
ZNRF3
|
zinc and ring finger 3 |
| chr20_+_35234223 | 0.11 |
ENST00000342422.3
|
C20orf24
|
chromosome 20 open reading frame 24 |
| chr20_+_48429233 | 0.11 |
ENST00000417961.1
|
SLC9A8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr7_-_74221288 | 0.11 |
ENST00000451013.2
|
GTF2IRD2
|
GTF2I repeat domain containing 2 |
| chr19_+_2785458 | 0.11 |
ENST00000307741.6
ENST00000585338.1 |
THOP1
|
thimet oligopeptidase 1 |
| chr22_+_37447771 | 0.10 |
ENST00000402077.3
ENST00000403888.3 ENST00000456470.1 |
KCTD17
|
potassium channel tetramerization domain containing 17 |
| chr3_-_52001448 | 0.10 |
ENST00000461554.1
ENST00000395013.3 ENST00000428823.2 ENST00000483411.1 ENST00000461544.1 ENST00000355852.2 |
PCBP4
|
poly(rC) binding protein 4 |
| chr19_+_18492973 | 0.10 |
ENST00000595973.2
|
GDF15
|
growth differentiation factor 15 |
| chr17_+_20483037 | 0.10 |
ENST00000399044.1
|
CDRT15L2
|
CMT1A duplicated region transcript 15-like 2 |
| chr2_+_47168313 | 0.10 |
ENST00000319190.5
ENST00000394850.2 ENST00000536057.1 |
TTC7A
|
tetratricopeptide repeat domain 7A |
| chr19_-_44258770 | 0.10 |
ENST00000601925.1
ENST00000602222.1 ENST00000599804.1 |
SMG9
|
SMG9 nonsense mediated mRNA decay factor |
| chr15_+_89164520 | 0.10 |
ENST00000332810.3
|
AEN
|
apoptosis enhancing nuclease |
| chr5_+_141348755 | 0.10 |
ENST00000506004.1
ENST00000507291.1 |
RNF14
|
ring finger protein 14 |
| chr11_+_36397528 | 0.10 |
ENST00000311599.5
ENST00000378867.3 |
PRR5L
|
proline rich 5 like |
| chr2_-_75788038 | 0.10 |
ENST00000393913.3
ENST00000410113.1 |
EVA1A
|
eva-1 homolog A (C. elegans) |
| chr19_+_42364313 | 0.10 |
ENST00000601492.1
ENST00000600467.1 ENST00000221975.2 |
RPS19
|
ribosomal protein S19 |
| chr19_-_42746714 | 0.10 |
ENST00000222330.3
|
GSK3A
|
glycogen synthase kinase 3 alpha |
| chr10_+_64564469 | 0.10 |
ENST00000373783.1
|
ADO
|
2-aminoethanethiol (cysteamine) dioxygenase |
| chr20_-_62680984 | 0.10 |
ENST00000340356.7
|
SOX18
|
SRY (sex determining region Y)-box 18 |
| chr15_-_89456630 | 0.10 |
ENST00000268150.8
|
MFGE8
|
milk fat globule-EGF factor 8 protein |
| chr2_-_242447732 | 0.10 |
ENST00000439101.1
ENST00000424537.1 ENST00000401869.1 ENST00000436402.1 |
STK25
|
serine/threonine kinase 25 |
| chr2_-_71221942 | 0.10 |
ENST00000272438.4
|
TEX261
|
testis expressed 261 |
| chr11_+_119038897 | 0.10 |
ENST00000454811.1
ENST00000449394.1 |
NLRX1
|
NLR family member X1 |
| chr19_-_40931891 | 0.10 |
ENST00000357949.4
|
SERTAD1
|
SERTA domain containing 1 |
| chr12_-_14996355 | 0.09 |
ENST00000228936.4
|
ART4
|
ADP-ribosyltransferase 4 (Dombrock blood group) |
| chr20_+_48429356 | 0.09 |
ENST00000361573.2
ENST00000541138.1 ENST00000539601.1 |
SLC9A8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr12_+_56546223 | 0.09 |
ENST00000550443.1
ENST00000207437.5 |
MYL6B
|
myosin, light chain 6B, alkali, smooth muscle and non-muscle |
| chr16_+_2079501 | 0.09 |
ENST00000563587.1
|
SLC9A3R2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chr5_-_146833803 | 0.09 |
ENST00000512722.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr3_+_183894566 | 0.09 |
ENST00000439647.1
|
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit |
| chr16_-_31105870 | 0.09 |
ENST00000394971.3
|
VKORC1
|
vitamin K epoxide reductase complex, subunit 1 |
| chr2_-_26205340 | 0.09 |
ENST00000264712.3
|
KIF3C
|
kinesin family member 3C |
| chr9_-_134406565 | 0.09 |
ENST00000372210.3
ENST00000372211.3 |
UCK1
|
uridine-cytidine kinase 1 |
| chr2_-_242447962 | 0.09 |
ENST00000405883.3
|
STK25
|
serine/threonine kinase 25 |
| chr17_+_18087553 | 0.09 |
ENST00000399138.4
|
ALKBH5
|
alkB, alkylation repair homolog 5 (E. coli) |
| chr19_-_44258733 | 0.09 |
ENST00000597586.1
ENST00000596714.1 |
SMG9
|
SMG9 nonsense mediated mRNA decay factor |
| chr19_+_42364460 | 0.09 |
ENST00000593863.1
|
RPS19
|
ribosomal protein S19 |
| chr12_-_59314246 | 0.08 |
ENST00000320743.3
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3 |
| chr1_-_6453399 | 0.08 |
ENST00000608083.1
|
ACOT7
|
acyl-CoA thioesterase 7 |
| chr2_-_242447983 | 0.08 |
ENST00000426941.1
ENST00000405585.1 ENST00000420551.1 ENST00000535007.1 ENST00000429279.1 ENST00000442307.1 ENST00000403346.3 ENST00000316586.4 |
STK25
|
serine/threonine kinase 25 |
| chr19_+_17982747 | 0.08 |
ENST00000222248.3
|
SLC5A5
|
solute carrier family 5 (sodium/iodide cotransporter), member 5 |
| chr19_+_926000 | 0.08 |
ENST00000263620.3
|
ARID3A
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr16_+_30207122 | 0.08 |
ENST00000395137.2
|
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr1_+_116519112 | 0.08 |
ENST00000369503.4
|
SLC22A15
|
solute carrier family 22, member 15 |
| chr19_+_827823 | 0.08 |
ENST00000233997.2
|
AZU1
|
azurocidin 1 |
| chr15_-_102285913 | 0.08 |
ENST00000558592.1
|
RP11-89K11.1
|
Uncharacterized protein |
| chr9_-_134406611 | 0.07 |
ENST00000372208.3
ENST00000372215.4 |
UCK1
|
uridine-cytidine kinase 1 |
| chr3_+_184279566 | 0.07 |
ENST00000330394.2
|
EPHB3
|
EPH receptor B3 |
| chrX_-_70473957 | 0.07 |
ENST00000373984.3
ENST00000314425.5 ENST00000373982.1 |
ZMYM3
|
zinc finger, MYM-type 3 |
| chr5_-_72861175 | 0.07 |
ENST00000504641.1
|
ANKRA2
|
ankyrin repeat, family A (RFXANK-like), 2 |
| chr16_+_29467780 | 0.07 |
ENST00000395400.3
|
SULT1A4
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 4 |
| chr17_+_12859080 | 0.07 |
ENST00000583608.1
|
ARHGAP44
|
Rho GTPase activating protein 44 |
| chr10_-_102279586 | 0.07 |
ENST00000370345.3
ENST00000451524.1 ENST00000370329.5 |
SEC31B
|
SEC31 homolog B (S. cerevisiae) |
| chr11_+_64002292 | 0.07 |
ENST00000426086.2
|
VEGFB
|
vascular endothelial growth factor B |
| chrX_-_153583257 | 0.07 |
ENST00000438732.1
|
FLNA
|
filamin A, alpha |
| chr11_+_65222698 | 0.06 |
ENST00000309775.7
|
AP000769.1
|
Uncharacterized protein |
| chr19_-_2456922 | 0.06 |
ENST00000582871.1
ENST00000325327.3 |
LMNB2
|
lamin B2 |
| chr1_-_45805752 | 0.06 |
ENST00000354383.6
ENST00000355498.2 ENST00000372100.5 ENST00000531105.1 |
MUTYH
|
mutY homolog |
| chr11_-_129062093 | 0.06 |
ENST00000310343.9
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr7_+_72742178 | 0.06 |
ENST00000442793.1
ENST00000413573.2 ENST00000252037.4 |
FKBP6
|
FK506 binding protein 6, 36kDa |
| chr1_+_25598872 | 0.06 |
ENST00000328664.4
|
RHD
|
Rh blood group, D antigen |
| chr3_-_52479043 | 0.06 |
ENST00000231721.2
ENST00000475739.1 |
SEMA3G
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3G |
| chr4_-_10118348 | 0.06 |
ENST00000502702.1
|
WDR1
|
WD repeat domain 1 |
| chr3_-_184079382 | 0.06 |
ENST00000344937.7
ENST00000423355.2 ENST00000434054.2 ENST00000457512.1 ENST00000265593.4 |
CLCN2
|
chloride channel, voltage-sensitive 2 |
| chr7_+_100770328 | 0.06 |
ENST00000223095.4
ENST00000445463.2 |
SERPINE1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
| chr8_-_22926526 | 0.06 |
ENST00000347739.3
ENST00000542226.1 |
TNFRSF10B
|
tumor necrosis factor receptor superfamily, member 10b |
Network of associatons between targets according to the STRING database.
First level regulatory network of TP53
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.1 | 0.3 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.1 | 0.6 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 0.5 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.1 | 0.2 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.1 | 0.3 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.5 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.1 | 0.2 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.1 | 0.2 | GO:1902512 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) positive regulation of apoptotic DNA fragmentation(GO:1902512) |
| 0.1 | 0.3 | GO:0051541 | negative regulation of extracellular matrix disassembly(GO:0010716) elastin metabolic process(GO:0051541) |
| 0.0 | 0.8 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 0.5 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.0 | 0.3 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:0071879 | UDP-glucose catabolic process(GO:0006258) positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.1 | GO:0045083 | negative regulation of interleukin-12 biosynthetic process(GO:0045083) |
| 0.0 | 0.2 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.0 | 0.2 | GO:2000771 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.1 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.0 | 0.6 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.1 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.0 | 0.6 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.3 | GO:0001767 | establishment of lymphocyte polarity(GO:0001767) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.1 | GO:1900535 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.0 | 0.1 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.0 | 0.5 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.3 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.3 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.0 | 0.2 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.1 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.0 | 0.1 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.0 | 0.3 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.0 | 0.3 | GO:0036481 | intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036481) |
| 0.0 | 0.1 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.2 | GO:0060315 | protein maturation by protein folding(GO:0022417) negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.2 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.2 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.0 | GO:0003095 | pressure natriuresis(GO:0003095) vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.3 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.2 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.2 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.1 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.1 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.2 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.0 | 0.3 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.5 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.3 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.3 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.1 | GO:0030120 | ER to Golgi transport vesicle membrane(GO:0012507) vesicle coat(GO:0030120) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.9 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.2 | 0.7 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.2 | 0.8 | GO:0004945 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.2 | 0.5 | GO:0052894 | norspermine:oxygen oxidoreductase activity(GO:0052894) N1-acetylspermine:oxygen oxidoreductase (N1-acetylspermidine-forming) activity(GO:0052895) |
| 0.1 | 0.4 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.1 | 0.4 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.1 | 0.5 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.0 | 0.5 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.3 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.3 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.3 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.4 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.2 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.2 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.2 | GO:0098634 | protein binding involved in cell-matrix adhesion(GO:0098634) |
| 0.0 | 0.1 | GO:0042835 | BRE binding(GO:0042835) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.1 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.2 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.9 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.0 | 0.2 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.5 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.1 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.6 | GO:0008536 | Ran GTPase binding(GO:0008536) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.2 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.5 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.5 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.5 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.2 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.7 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |