Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
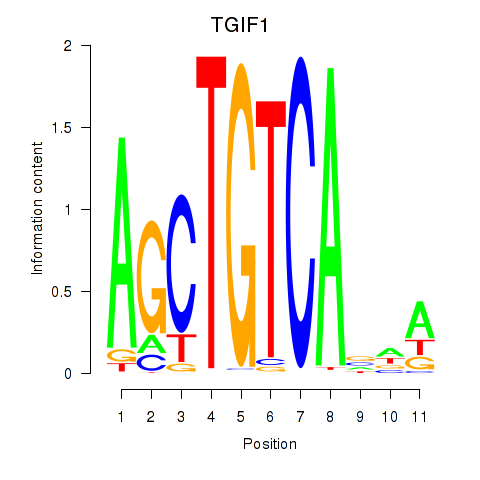
Results for TGIF1
Z-value: 0.48
Transcription factors associated with TGIF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TGIF1
|
ENSG00000177426.16 | TGFB induced factor homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TGIF1 | hg19_v2_chr18_+_3447572_3447607 | -1.00 | 1.2e-03 | Click! |
Activity profile of TGIF1 motif
Sorted Z-values of TGIF1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_65337708 | 0.27 |
ENST00000334287.2
|
SLC51B
|
solute carrier family 51, beta subunit |
| chr8_+_72755796 | 0.26 |
ENST00000524152.1
|
RP11-383H13.1
|
Protein LOC100132891; cDNA FLJ53548 |
| chr11_+_71934962 | 0.26 |
ENST00000543234.1
|
INPPL1
|
inositol polyphosphate phosphatase-like 1 |
| chr10_+_86004802 | 0.24 |
ENST00000359452.4
ENST00000358110.5 ENST00000372092.3 |
RGR
|
retinal G protein coupled receptor |
| chr7_+_26438187 | 0.22 |
ENST00000439120.1
ENST00000430548.1 ENST00000421862.1 ENST00000449537.1 ENST00000420774.1 ENST00000418758.2 |
AC004540.5
|
AC004540.5 |
| chr3_-_52719912 | 0.22 |
ENST00000420148.1
|
PBRM1
|
polybromo 1 |
| chr16_+_88636875 | 0.21 |
ENST00000569435.1
|
ZC3H18
|
zinc finger CCCH-type containing 18 |
| chr12_-_62653903 | 0.19 |
ENST00000552075.1
|
FAM19A2
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2 |
| chr12_-_71512027 | 0.19 |
ENST00000549357.1
|
CTD-2021H9.3
|
Uncharacterized protein |
| chr2_-_3595547 | 0.19 |
ENST00000438485.1
|
RP13-512J5.1
|
Uncharacterized protein |
| chr2_+_128177458 | 0.18 |
ENST00000409048.1
ENST00000422777.3 |
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr3_+_187461442 | 0.18 |
ENST00000450760.1
|
RP11-211G3.2
|
RP11-211G3.2 |
| chr16_+_2880254 | 0.18 |
ENST00000570670.1
|
ZG16B
|
zymogen granule protein 16B |
| chr8_+_40018977 | 0.18 |
ENST00000520487.1
|
RP11-470M17.2
|
RP11-470M17.2 |
| chr2_+_37311588 | 0.17 |
ENST00000409774.1
ENST00000608836.1 |
GPATCH11
|
G patch domain containing 11 |
| chr19_-_47164386 | 0.17 |
ENST00000391916.2
ENST00000410105.2 |
DACT3
|
dishevelled-binding antagonist of beta-catenin 3 |
| chr1_-_54872059 | 0.16 |
ENST00000371320.3
|
SSBP3
|
single stranded DNA binding protein 3 |
| chr5_+_149865377 | 0.16 |
ENST00000522491.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr8_-_29120604 | 0.16 |
ENST00000521515.1
|
KIF13B
|
kinesin family member 13B |
| chr12_+_95611536 | 0.16 |
ENST00000549002.1
|
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr3_-_52719810 | 0.16 |
ENST00000424867.1
ENST00000394830.3 ENST00000431678.1 ENST00000450271.1 |
PBRM1
|
polybromo 1 |
| chr4_+_108815402 | 0.16 |
ENST00000503385.1
|
SGMS2
|
sphingomyelin synthase 2 |
| chr14_-_75083313 | 0.15 |
ENST00000556652.1
ENST00000555313.1 |
CTD-2207P18.2
|
CTD-2207P18.2 |
| chr7_-_50628745 | 0.15 |
ENST00000380984.4
ENST00000357936.5 ENST00000426377.1 |
DDC
|
dopa decarboxylase (aromatic L-amino acid decarboxylase) |
| chr20_+_4667094 | 0.15 |
ENST00000424424.1
ENST00000457586.1 |
PRNP
|
prion protein |
| chr1_-_91317072 | 0.14 |
ENST00000435649.2
ENST00000443802.1 |
RP4-665J23.1
|
RP4-665J23.1 |
| chr8_+_38239882 | 0.14 |
ENST00000607047.1
|
RP11-350N15.5
|
RP11-350N15.5 |
| chr6_-_72129806 | 0.14 |
ENST00000413945.1
ENST00000602878.1 ENST00000436803.1 ENST00000421704.1 ENST00000441570.1 |
LINC00472
|
long intergenic non-protein coding RNA 472 |
| chr3_+_63897605 | 0.14 |
ENST00000487717.1
|
ATXN7
|
ataxin 7 |
| chr11_+_64052294 | 0.14 |
ENST00000536667.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr22_-_36013368 | 0.13 |
ENST00000442617.1
ENST00000397326.2 ENST00000397328.1 ENST00000451685.1 |
MB
|
myoglobin |
| chr5_-_130868688 | 0.13 |
ENST00000504575.1
ENST00000513227.1 |
RAPGEF6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr5_-_74348371 | 0.12 |
ENST00000503568.1
|
RP11-229C3.2
|
RP11-229C3.2 |
| chr20_-_35807741 | 0.12 |
ENST00000434295.1
ENST00000441008.2 ENST00000400441.3 ENST00000343811.4 |
MROH8
|
maestro heat-like repeat family member 8 |
| chr20_-_50418947 | 0.12 |
ENST00000371539.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr1_-_89357179 | 0.12 |
ENST00000448623.1
ENST00000418217.1 ENST00000370500.5 |
GTF2B
|
general transcription factor IIB |
| chr4_+_186990298 | 0.12 |
ENST00000296795.3
ENST00000513189.1 |
TLR3
|
toll-like receptor 3 |
| chr14_-_23762777 | 0.11 |
ENST00000431326.2
|
HOMEZ
|
homeobox and leucine zipper encoding |
| chr4_+_128886584 | 0.11 |
ENST00000513371.1
|
C4orf29
|
chromosome 4 open reading frame 29 |
| chr2_+_242127924 | 0.11 |
ENST00000402530.3
ENST00000274979.8 ENST00000402430.3 |
ANO7
|
anoctamin 7 |
| chr4_-_83483094 | 0.11 |
ENST00000508701.1
ENST00000454948.3 |
TMEM150C
|
transmembrane protein 150C |
| chr2_+_97202480 | 0.11 |
ENST00000357485.3
|
ARID5A
|
AT rich interactive domain 5A (MRF1-like) |
| chr12_+_95612006 | 0.10 |
ENST00000551311.1
ENST00000546445.1 |
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr1_-_150978953 | 0.10 |
ENST00000493834.2
|
FAM63A
|
family with sequence similarity 63, member A |
| chr12_+_79371565 | 0.10 |
ENST00000551304.1
|
SYT1
|
synaptotagmin I |
| chr11_+_64052454 | 0.10 |
ENST00000539833.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr1_+_95616933 | 0.10 |
ENST00000604203.1
|
RP11-57H12.6
|
TMEM56-RWDD3 readthrough |
| chr5_-_58571935 | 0.10 |
ENST00000503258.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr9_+_37079968 | 0.10 |
ENST00000588403.1
|
RP11-220I1.1
|
RP11-220I1.1 |
| chr1_+_150039877 | 0.09 |
ENST00000419023.1
|
VPS45
|
vacuolar protein sorting 45 homolog (S. cerevisiae) |
| chr11_+_64052266 | 0.09 |
ENST00000539851.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr4_+_14113592 | 0.09 |
ENST00000502759.1
ENST00000511200.1 ENST00000512754.1 ENST00000506739.1 |
LINC01085
|
long intergenic non-protein coding RNA 1085 |
| chr17_-_71223839 | 0.09 |
ENST00000579872.1
ENST00000580032.1 |
FAM104A
|
family with sequence similarity 104, member A |
| chr3_+_122920847 | 0.09 |
ENST00000466519.1
ENST00000480631.1 ENST00000491366.1 ENST00000487572.1 |
SEC22A
|
SEC22 vesicle trafficking protein homolog A (S. cerevisiae) |
| chr15_-_42840961 | 0.09 |
ENST00000563454.1
ENST00000397130.3 ENST00000570160.1 ENST00000323443.2 |
LRRC57
|
leucine rich repeat containing 57 |
| chr14_+_31494672 | 0.09 |
ENST00000542754.2
ENST00000313566.6 |
AP4S1
|
adaptor-related protein complex 4, sigma 1 subunit |
| chr1_-_155959280 | 0.09 |
ENST00000495070.2
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
| chr16_-_67217844 | 0.09 |
ENST00000563902.1
ENST00000561621.1 ENST00000290881.7 |
KIAA0895L
|
KIAA0895-like |
| chr2_-_216003127 | 0.09 |
ENST00000412081.1
ENST00000272895.7 |
ABCA12
|
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr2_+_39005325 | 0.09 |
ENST00000281950.3
|
GEMIN6
|
gem (nuclear organelle) associated protein 6 |
| chr1_-_150979333 | 0.09 |
ENST00000312210.5
|
FAM63A
|
family with sequence similarity 63, member A |
| chr8_-_29120580 | 0.09 |
ENST00000524189.1
|
KIF13B
|
kinesin family member 13B |
| chr22_+_20905422 | 0.09 |
ENST00000424287.1
ENST00000423862.1 |
MED15
|
mediator complex subunit 15 |
| chr1_+_225600404 | 0.09 |
ENST00000366845.2
|
AC092811.1
|
AC092811.1 |
| chr14_+_70078303 | 0.09 |
ENST00000342745.4
|
KIAA0247
|
KIAA0247 |
| chr1_+_150039787 | 0.09 |
ENST00000535106.1
|
VPS45
|
vacuolar protein sorting 45 homolog (S. cerevisiae) |
| chr6_-_72130472 | 0.09 |
ENST00000426635.2
|
LINC00472
|
long intergenic non-protein coding RNA 472 |
| chr17_-_34122596 | 0.09 |
ENST00000250144.8
|
MMP28
|
matrix metallopeptidase 28 |
| chr5_-_65018834 | 0.09 |
ENST00000506816.1
|
SGTB
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr20_-_50418972 | 0.08 |
ENST00000395997.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr5_+_76012009 | 0.08 |
ENST00000505600.1
|
F2R
|
coagulation factor II (thrombin) receptor |
| chr9_+_37120536 | 0.08 |
ENST00000336755.5
ENST00000534928.1 ENST00000322831.6 |
ZCCHC7
|
zinc finger, CCHC domain containing 7 |
| chr2_-_36779411 | 0.08 |
ENST00000406220.1
|
AC007401.2
|
Uncharacterized protein |
| chr15_-_78112553 | 0.08 |
ENST00000562933.1
|
LINGO1
|
leucine rich repeat and Ig domain containing 1 |
| chr8_-_74495065 | 0.08 |
ENST00000523533.1
|
STAU2
|
staufen double-stranded RNA binding protein 2 |
| chr8_+_38243721 | 0.08 |
ENST00000527334.1
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr15_-_55562451 | 0.08 |
ENST00000568803.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr3_-_127541194 | 0.08 |
ENST00000453507.2
|
MGLL
|
monoglyceride lipase |
| chr9_+_2159850 | 0.08 |
ENST00000416751.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr8_+_99076509 | 0.08 |
ENST00000318528.3
|
C8orf47
|
chromosome 8 open reading frame 47 |
| chr14_-_23299009 | 0.08 |
ENST00000488800.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr3_+_160473343 | 0.08 |
ENST00000497343.1
|
PPM1L
|
protein phosphatase, Mg2+/Mn2+ dependent, 1L |
| chr7_+_120591170 | 0.08 |
ENST00000431467.1
|
ING3
|
inhibitor of growth family, member 3 |
| chr13_-_34250861 | 0.08 |
ENST00000445227.1
ENST00000454681.2 |
RP11-141M1.3
|
RP11-141M1.3 |
| chr1_-_161046266 | 0.07 |
ENST00000453926.2
|
PVRL4
|
poliovirus receptor-related 4 |
| chr9_-_132805430 | 0.07 |
ENST00000446176.2
ENST00000355681.3 ENST00000420781.1 |
FNBP1
|
formin binding protein 1 |
| chr17_+_8316442 | 0.07 |
ENST00000582812.1
|
NDEL1
|
nudE neurodevelopment protein 1-like 1 |
| chr1_+_205473720 | 0.07 |
ENST00000429964.2
ENST00000506784.1 ENST00000360066.2 |
CDK18
|
cyclin-dependent kinase 18 |
| chr4_+_8183793 | 0.07 |
ENST00000509119.1
|
SH3TC1
|
SH3 domain and tetratricopeptide repeats 1 |
| chrX_+_47696337 | 0.07 |
ENST00000334937.4
ENST00000376950.4 |
ZNF81
|
zinc finger protein 81 |
| chr12_+_25348186 | 0.07 |
ENST00000555711.1
ENST00000556885.1 ENST00000554266.1 ENST00000556351.1 ENST00000556927.1 ENST00000556402.1 ENST00000553788.1 |
LYRM5
|
LYR motif containing 5 |
| chr4_+_24661479 | 0.07 |
ENST00000569621.1
|
RP11-496D24.2
|
RP11-496D24.2 |
| chr5_-_132112921 | 0.07 |
ENST00000378721.4
ENST00000378701.1 |
SEPT8
|
septin 8 |
| chr12_-_46121554 | 0.07 |
ENST00000609803.1
|
LINC00938
|
long intergenic non-protein coding RNA 938 |
| chrX_-_128788914 | 0.07 |
ENST00000429967.1
ENST00000307484.6 |
APLN
|
apelin |
| chr5_-_168006324 | 0.07 |
ENST00000522176.1
|
PANK3
|
pantothenate kinase 3 |
| chr15_-_70994612 | 0.07 |
ENST00000558758.1
ENST00000379983.2 ENST00000560441.1 |
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr2_+_85804614 | 0.07 |
ENST00000263864.5
ENST00000409760.1 |
VAMP8
|
vesicle-associated membrane protein 8 |
| chr11_-_8954491 | 0.07 |
ENST00000526227.1
ENST00000525780.1 ENST00000326053.5 |
C11orf16
|
chromosome 11 open reading frame 16 |
| chr2_+_90458201 | 0.07 |
ENST00000603238.1
|
CH17-132F21.1
|
Uncharacterized protein |
| chr12_+_25348139 | 0.06 |
ENST00000557540.2
ENST00000381356.4 |
LYRM5
|
LYR motif containing 5 |
| chr1_+_110577229 | 0.06 |
ENST00000369795.3
ENST00000369794.2 |
STRIP1
|
striatin interacting protein 1 |
| chr4_-_175443788 | 0.06 |
ENST00000541923.1
|
HPGD
|
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr5_+_40841410 | 0.06 |
ENST00000381677.3
|
CARD6
|
caspase recruitment domain family, member 6 |
| chr2_+_136343943 | 0.06 |
ENST00000409606.1
|
R3HDM1
|
R3H domain containing 1 |
| chr17_-_57232596 | 0.06 |
ENST00000581068.1
ENST00000330137.7 |
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr19_+_7953417 | 0.06 |
ENST00000600345.1
ENST00000598224.1 |
LRRC8E
|
leucine rich repeat containing 8 family, member E |
| chr2_+_39005336 | 0.06 |
ENST00000409566.1
|
GEMIN6
|
gem (nuclear organelle) associated protein 6 |
| chr17_-_42188598 | 0.06 |
ENST00000591714.1
|
HDAC5
|
histone deacetylase 5 |
| chr2_-_74780176 | 0.06 |
ENST00000409549.1
|
LOXL3
|
lysyl oxidase-like 3 |
| chr4_-_46391367 | 0.06 |
ENST00000503806.1
ENST00000356504.1 ENST00000514090.1 ENST00000506961.1 |
GABRA2
|
gamma-aminobutyric acid (GABA) A receptor, alpha 2 |
| chr8_-_41522779 | 0.06 |
ENST00000522231.1
ENST00000314214.8 ENST00000348036.4 ENST00000457297.1 ENST00000522543.1 |
ANK1
|
ankyrin 1, erythrocytic |
| chr2_+_27505260 | 0.06 |
ENST00000380075.2
ENST00000296098.4 |
TRIM54
|
tripartite motif containing 54 |
| chr9_-_134615326 | 0.06 |
ENST00000438647.1
|
RAPGEF1
|
Rap guanine nucleotide exchange factor (GEF) 1 |
| chr13_-_48575376 | 0.06 |
ENST00000434484.1
|
SUCLA2
|
succinate-CoA ligase, ADP-forming, beta subunit |
| chr16_+_2588012 | 0.06 |
ENST00000354836.5
ENST00000389224.3 |
PDPK1
|
3-phosphoinositide dependent protein kinase-1 |
| chr2_+_97203082 | 0.06 |
ENST00000454558.2
|
ARID5A
|
AT rich interactive domain 5A (MRF1-like) |
| chr19_+_49375649 | 0.06 |
ENST00000200453.5
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A |
| chr7_+_95401877 | 0.06 |
ENST00000524053.1
ENST00000324972.6 ENST00000537881.1 ENST00000437599.1 ENST00000359388.4 ENST00000413338.1 |
DYNC1I1
|
dynein, cytoplasmic 1, intermediate chain 1 |
| chr17_+_57233087 | 0.06 |
ENST00000578777.1
ENST00000577457.1 ENST00000582995.1 |
PRR11
|
proline rich 11 |
| chr22_+_37959647 | 0.06 |
ENST00000415670.1
|
CDC42EP1
|
CDC42 effector protein (Rho GTPase binding) 1 |
| chr15_+_40733387 | 0.06 |
ENST00000416165.1
|
BAHD1
|
bromo adjacent homology domain containing 1 |
| chr3_+_185080908 | 0.06 |
ENST00000265026.3
|
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr2_+_33661382 | 0.06 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr22_+_35653445 | 0.06 |
ENST00000420166.1
ENST00000444518.2 ENST00000455359.1 ENST00000216106.5 |
HMGXB4
|
HMG box domain containing 4 |
| chr10_+_94608218 | 0.06 |
ENST00000371543.1
|
EXOC6
|
exocyst complex component 6 |
| chrX_-_80065146 | 0.06 |
ENST00000373275.4
|
BRWD3
|
bromodomain and WD repeat domain containing 3 |
| chr7_+_23637763 | 0.06 |
ENST00000410069.1
|
CCDC126
|
coiled-coil domain containing 126 |
| chr14_+_31494841 | 0.06 |
ENST00000556232.1
ENST00000216366.4 ENST00000334725.4 ENST00000554609.1 ENST00000554345.1 |
AP4S1
|
adaptor-related protein complex 4, sigma 1 subunit |
| chr16_+_88636789 | 0.06 |
ENST00000301011.5
ENST00000452588.2 |
ZC3H18
|
zinc finger CCCH-type containing 18 |
| chr5_+_141348755 | 0.06 |
ENST00000506004.1
ENST00000507291.1 |
RNF14
|
ring finger protein 14 |
| chr3_+_156807663 | 0.06 |
ENST00000467995.1
ENST00000474477.1 ENST00000471719.1 |
LINC00881
|
long intergenic non-protein coding RNA 881 |
| chr8_+_67039278 | 0.05 |
ENST00000276573.7
ENST00000350034.4 |
TRIM55
|
tripartite motif containing 55 |
| chr2_-_113594279 | 0.05 |
ENST00000416750.1
ENST00000418817.1 ENST00000263341.2 |
IL1B
|
interleukin 1, beta |
| chr1_+_111888890 | 0.05 |
ENST00000369738.4
|
PIFO
|
primary cilia formation |
| chr6_-_138866823 | 0.05 |
ENST00000342260.5
|
NHSL1
|
NHS-like 1 |
| chr5_-_132113063 | 0.05 |
ENST00000378719.2
|
SEPT8
|
septin 8 |
| chr19_+_10947251 | 0.05 |
ENST00000592854.1
|
C19orf38
|
chromosome 19 open reading frame 38 |
| chr8_+_38243821 | 0.05 |
ENST00000519476.2
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr9_-_97356075 | 0.05 |
ENST00000375337.3
|
FBP2
|
fructose-1,6-bisphosphatase 2 |
| chr8_-_67090825 | 0.05 |
ENST00000276571.3
|
CRH
|
corticotropin releasing hormone |
| chr10_+_94608245 | 0.05 |
ENST00000443748.2
ENST00000260762.6 |
EXOC6
|
exocyst complex component 6 |
| chr17_-_57232525 | 0.05 |
ENST00000583380.1
ENST00000580541.1 ENST00000578105.1 ENST00000437036.2 |
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr20_-_50419055 | 0.05 |
ENST00000217086.4
|
SALL4
|
spalt-like transcription factor 4 |
| chr3_-_112280709 | 0.05 |
ENST00000402314.2
ENST00000283290.5 ENST00000492886.1 |
ATG3
|
autophagy related 3 |
| chr15_+_42841008 | 0.05 |
ENST00000260372.3
ENST00000568876.1 ENST00000568846.2 ENST00000562398.1 |
HAUS2
|
HAUS augmin-like complex, subunit 2 |
| chr5_-_132112907 | 0.05 |
ENST00000458488.2
|
SEPT8
|
septin 8 |
| chr3_+_107318157 | 0.05 |
ENST00000406780.1
|
BBX
|
bobby sox homolog (Drosophila) |
| chr4_+_55095428 | 0.05 |
ENST00000508170.1
ENST00000512143.1 |
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr8_+_38244638 | 0.05 |
ENST00000526356.1
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr5_-_127873896 | 0.05 |
ENST00000502468.1
|
FBN2
|
fibrillin 2 |
| chr1_-_33168336 | 0.05 |
ENST00000373484.3
|
SYNC
|
syncoilin, intermediate filament protein |
| chr7_-_107770794 | 0.05 |
ENST00000205386.4
ENST00000418464.1 ENST00000388781.3 ENST00000388780.3 ENST00000414450.2 |
LAMB4
|
laminin, beta 4 |
| chr4_-_122085469 | 0.05 |
ENST00000057513.3
|
TNIP3
|
TNFAIP3 interacting protein 3 |
| chr4_+_95972822 | 0.05 |
ENST00000509540.1
ENST00000440890.2 |
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr9_+_128510454 | 0.05 |
ENST00000491787.3
ENST00000447726.2 |
PBX3
|
pre-B-cell leukemia homeobox 3 |
| chr20_+_35201993 | 0.05 |
ENST00000373872.4
|
TGIF2
|
TGFB-induced factor homeobox 2 |
| chr17_+_74734052 | 0.05 |
ENST00000590514.1
|
MFSD11
|
major facilitator superfamily domain containing 11 |
| chr5_+_180257974 | 0.05 |
ENST00000502162.2
ENST00000508309.1 ENST00000509921.1 |
LINC00847
|
long intergenic non-protein coding RNA 847 |
| chr16_+_31044812 | 0.05 |
ENST00000313843.3
|
STX4
|
syntaxin 4 |
| chr6_+_80714318 | 0.05 |
ENST00000369798.2
|
TTK
|
TTK protein kinase |
| chr2_+_37311645 | 0.05 |
ENST00000281932.5
|
GPATCH11
|
G patch domain containing 11 |
| chr16_-_30582888 | 0.05 |
ENST00000563707.1
ENST00000567855.1 |
ZNF688
|
zinc finger protein 688 |
| chr20_+_43029911 | 0.05 |
ENST00000443598.2
ENST00000316099.4 ENST00000415691.2 |
HNF4A
|
hepatocyte nuclear factor 4, alpha |
| chr11_-_8795787 | 0.05 |
ENST00000528196.1
ENST00000533681.1 |
ST5
|
suppression of tumorigenicity 5 |
| chr22_-_37882395 | 0.05 |
ENST00000416983.3
ENST00000424765.2 ENST00000356998.3 |
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr4_-_2420357 | 0.05 |
ENST00000511071.1
ENST00000509171.1 ENST00000290974.2 |
ZFYVE28
|
zinc finger, FYVE domain containing 28 |
| chr2_+_242089833 | 0.05 |
ENST00000404405.3
ENST00000439916.1 ENST00000406106.3 ENST00000401987.1 |
PPP1R7
|
protein phosphatase 1, regulatory subunit 7 |
| chr9_-_35812236 | 0.05 |
ENST00000340291.2
|
SPAG8
|
sperm associated antigen 8 |
| chr5_+_43192311 | 0.05 |
ENST00000326035.2
|
NIM1
|
NIM1 serine/threonine protein kinase |
| chr6_-_111888474 | 0.05 |
ENST00000368735.1
|
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr19_-_45826125 | 0.05 |
ENST00000221476.3
|
CKM
|
creatine kinase, muscle |
| chr11_-_111649015 | 0.05 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr22_-_39150947 | 0.05 |
ENST00000411587.2
ENST00000420859.1 ENST00000452294.1 ENST00000456894.1 |
SUN2
|
Sad1 and UNC84 domain containing 2 |
| chr6_+_17600576 | 0.05 |
ENST00000259963.3
|
FAM8A1
|
family with sequence similarity 8, member A1 |
| chr4_+_146402843 | 0.05 |
ENST00000514831.1
|
SMAD1
|
SMAD family member 1 |
| chr2_+_136343820 | 0.05 |
ENST00000410054.1
|
R3HDM1
|
R3H domain containing 1 |
| chr10_+_28822417 | 0.05 |
ENST00000428935.1
ENST00000420266.1 |
WAC
|
WW domain containing adaptor with coiled-coil |
| chr9_+_4662282 | 0.04 |
ENST00000381883.2
|
PPAPDC2
|
phosphatidic acid phosphatase type 2 domain containing 2 |
| chr3_+_4535155 | 0.04 |
ENST00000544951.1
|
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr3_+_169629354 | 0.04 |
ENST00000428432.2
ENST00000335556.3 |
SAMD7
|
sterile alpha motif domain containing 7 |
| chr18_+_54318566 | 0.04 |
ENST00000589935.1
ENST00000357574.3 |
WDR7
|
WD repeat domain 7 |
| chr11_-_82745238 | 0.04 |
ENST00000531021.1
|
RAB30
|
RAB30, member RAS oncogene family |
| chr11_-_842509 | 0.04 |
ENST00000322028.4
|
POLR2L
|
polymerase (RNA) II (DNA directed) polypeptide L, 7.6kDa |
| chr6_+_109416684 | 0.04 |
ENST00000521522.1
ENST00000524064.1 ENST00000522608.1 ENST00000521503.1 ENST00000519407.1 ENST00000519095.1 ENST00000368968.2 ENST00000522490.1 ENST00000523209.1 ENST00000368970.2 ENST00000520883.1 ENST00000523787.1 |
CEP57L1
|
centrosomal protein 57kDa-like 1 |
| chr4_-_175443484 | 0.04 |
ENST00000514584.1
ENST00000542498.1 ENST00000296521.7 ENST00000422112.2 ENST00000504433.1 |
HPGD
|
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr5_-_179047881 | 0.04 |
ENST00000521173.1
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr7_-_97501469 | 0.04 |
ENST00000414884.1
ENST00000442734.1 |
ASNS
|
asparagine synthetase (glutamine-hydrolyzing) |
| chr7_+_134551583 | 0.04 |
ENST00000435928.1
|
CALD1
|
caldesmon 1 |
| chr9_-_35812140 | 0.04 |
ENST00000497810.1
ENST00000396638.2 ENST00000484764.1 |
SPAG8
|
sperm associated antigen 8 |
| chrX_-_99987088 | 0.04 |
ENST00000372981.1
ENST00000263033.5 |
SYTL4
|
synaptotagmin-like 4 |
| chr3_+_52719936 | 0.04 |
ENST00000418458.1
ENST00000394799.2 |
GNL3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr3_-_123168551 | 0.04 |
ENST00000462833.1
|
ADCY5
|
adenylate cyclase 5 |
| chr13_-_96329048 | 0.04 |
ENST00000606011.1
ENST00000499499.2 |
DNAJC3-AS1
|
DNAJC3 antisense RNA 1 (head to head) |
| chr15_-_43559055 | 0.04 |
ENST00000220420.5
ENST00000349114.4 |
TGM5
|
transglutaminase 5 |
| chr17_-_26220366 | 0.04 |
ENST00000460380.2
ENST00000508862.1 ENST00000379102.3 ENST00000582441.1 |
LYRM9
RP1-66C13.4
|
LYR motif containing 9 Uncharacterized protein |
| chr4_+_77870960 | 0.04 |
ENST00000505788.1
ENST00000510515.1 ENST00000504637.1 |
SEPT11
|
septin 11 |
| chr1_+_186649754 | 0.04 |
ENST00000608917.1
|
RP5-973M2.2
|
RP5-973M2.2 |
| chr1_+_221051699 | 0.04 |
ENST00000366903.6
|
HLX
|
H2.0-like homeobox |
| chr2_-_18770812 | 0.04 |
ENST00000359846.2
ENST00000304081.4 ENST00000600945.1 ENST00000532967.1 ENST00000444297.2 |
NT5C1B
NT5C1B-RDH14
|
5'-nucleotidase, cytosolic IB NT5C1B-RDH14 readthrough |
Network of associatons between targets according to the STRING database.
First level regulatory network of TGIF1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0097065 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.1 | 0.2 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.0 | 0.1 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) isoquinoline alkaloid metabolic process(GO:0033076) |
| 0.0 | 0.1 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.1 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.0 | 0.1 | GO:1901993 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.1 | GO:0043309 | regulation of eosinophil degranulation(GO:0043309) positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0043396 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.3 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.0 | 0.1 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.1 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.0 | 0.1 | GO:1900133 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) regulation of renin secretion into blood stream(GO:1900133) |
| 0.0 | 0.1 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.0 | 0.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.2 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.1 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.0 | 0.0 | GO:0007388 | anterior compartment pattern formation(GO:0007387) posterior compartment specification(GO:0007388) |
| 0.0 | 0.1 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 | 0.1 | GO:0042322 | negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) |
| 0.0 | 0.1 | GO:0043353 | slow-twitch skeletal muscle fiber contraction(GO:0031444) enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.0 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.1 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 0.0 | 0.1 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.0 | 0.1 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.1 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.0 | 0.2 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.0 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.0 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.0 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 | 0.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.1 | GO:0072277 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.0 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.0 | 0.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.1 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.1 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.1 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.5 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 0.2 | GO:0033270 | paranode region of axon(GO:0033270) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.0 | 0.1 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.0 | 0.2 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.2 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.1 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.0 | 0.1 | GO:0000384 | first spliceosomal transesterification activity(GO:0000384) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.0 | 0.1 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.0 | 0.1 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 0.1 | GO:0005018 | platelet-derived growth factor alpha-receptor activity(GO:0005018) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.2 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |