Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
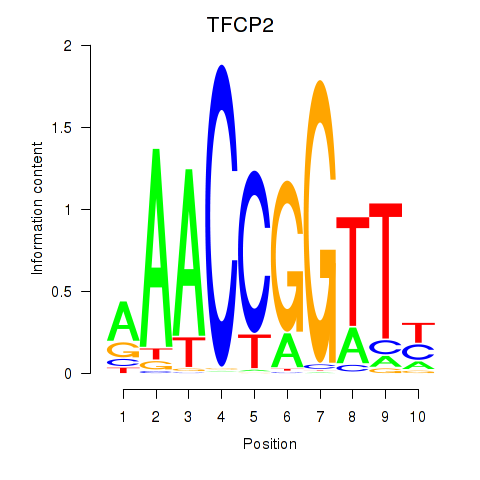
Results for TFCP2
Z-value: 0.98
Transcription factors associated with TFCP2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TFCP2
|
ENSG00000135457.5 | transcription factor CP2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TFCP2 | hg19_v2_chr12_-_51566562_51566584 | 0.67 | 3.3e-01 | Click! |
Activity profile of TFCP2 motif
Sorted Z-values of TFCP2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_17326521 | 0.42 |
ENST00000593597.1
|
USE1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr1_+_95975672 | 0.41 |
ENST00000440116.2
ENST00000456933.1 |
RP11-286B14.1
|
RP11-286B14.1 |
| chr19_-_57678811 | 0.32 |
ENST00000554048.2
|
DUXA
|
double homeobox A |
| chr4_-_174255400 | 0.31 |
ENST00000506267.1
|
HMGB2
|
high mobility group box 2 |
| chr12_+_93963590 | 0.31 |
ENST00000340600.2
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chr11_-_96123022 | 0.30 |
ENST00000542662.1
|
CCDC82
|
coiled-coil domain containing 82 |
| chr19_-_12146483 | 0.29 |
ENST00000455504.2
ENST00000547560.1 ENST00000552904.1 ENST00000419886.2 ENST00000550507.1 ENST00000344980.6 ENST00000550745.1 ENST00000411841.1 |
ZNF433
|
zinc finger protein 433 |
| chr16_-_28367975 | 0.28 |
ENST00000533640.1
|
NPIPB6
|
nuclear pore complex interacting protein family, member B6 |
| chr19_-_11849697 | 0.28 |
ENST00000586121.1
ENST00000431998.1 ENST00000341191.6 ENST00000545749.1 ENST00000440527.1 |
ZNF823
|
zinc finger protein 823 |
| chr4_-_174255536 | 0.28 |
ENST00000446922.2
|
HMGB2
|
high mobility group box 2 |
| chr19_+_46531127 | 0.27 |
ENST00000601033.1
|
CTC-344H19.4
|
CTC-344H19.4 |
| chr5_+_32712363 | 0.26 |
ENST00000507141.1
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr16_+_19098178 | 0.25 |
ENST00000568032.1
|
RP11-626G11.4
|
RP11-626G11.4 |
| chr16_+_74411776 | 0.24 |
ENST00000429990.1
|
NPIPB15
|
nuclear pore complex interacting protein family, member B15 |
| chr5_+_67584523 | 0.23 |
ENST00000521409.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr19_+_2841433 | 0.23 |
ENST00000334241.4
ENST00000585966.1 ENST00000591539.1 |
ZNF555
|
zinc finger protein 555 |
| chr21_-_40720974 | 0.23 |
ENST00000380748.1
|
HMGN1
|
high mobility group nucleosome binding domain 1 |
| chr18_+_56531584 | 0.23 |
ENST00000590287.1
|
ZNF532
|
zinc finger protein 532 |
| chr6_+_24777040 | 0.22 |
ENST00000378059.3
|
GMNN
|
geminin, DNA replication inhibitor |
| chr22_+_45809560 | 0.22 |
ENST00000342894.3
ENST00000538017.1 |
RIBC2
|
RIB43A domain with coiled-coils 2 |
| chr14_-_82089405 | 0.22 |
ENST00000554211.1
|
RP11-799P8.1
|
RP11-799P8.1 |
| chr17_-_57229155 | 0.22 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr10_-_90611566 | 0.22 |
ENST00000371930.4
|
ANKRD22
|
ankyrin repeat domain 22 |
| chr11_+_113185292 | 0.21 |
ENST00000429951.1
ENST00000442859.1 ENST00000531164.1 ENST00000529850.1 ENST00000314756.3 ENST00000525965.1 |
TTC12
|
tetratricopeptide repeat domain 12 |
| chrX_-_46759138 | 0.21 |
ENST00000377879.3
|
CXorf31
|
chromosome X open reading frame 31 |
| chr10_+_134232503 | 0.21 |
ENST00000428814.1
|
RP11-432J24.2
|
RP11-432J24.2 |
| chr5_+_136070614 | 0.20 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1 |
| chr1_-_145382434 | 0.20 |
ENST00000610154.1
|
RP11-458D21.1
|
RP11-458D21.1 |
| chr16_+_15032072 | 0.20 |
ENST00000424549.3
|
NPIPA1
|
nuclear pore complex interacting protein family, member A1 |
| chr18_+_20494078 | 0.20 |
ENST00000579124.1
ENST00000577588.1 ENST00000582354.1 ENST00000581819.1 |
RBBP8
|
retinoblastoma binding protein 8 |
| chr4_+_128702969 | 0.19 |
ENST00000508776.1
ENST00000439123.2 |
HSPA4L
|
heat shock 70kDa protein 4-like |
| chr1_+_16330723 | 0.19 |
ENST00000329454.2
|
C1orf64
|
chromosome 1 open reading frame 64 |
| chr16_+_20817746 | 0.19 |
ENST00000568894.1
|
AC004381.6
|
Putative RNA exonuclease NEF-sp |
| chr19_-_37019136 | 0.19 |
ENST00000592282.1
|
ZNF260
|
zinc finger protein 260 |
| chr2_+_189156389 | 0.19 |
ENST00000409843.1
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr5_-_132362226 | 0.18 |
ENST00000509437.1
ENST00000355372.2 ENST00000513541.1 ENST00000509008.1 ENST00000513848.1 ENST00000504170.1 ENST00000324170.3 |
ZCCHC10
|
zinc finger, CCHC domain containing 10 |
| chr18_-_54305658 | 0.18 |
ENST00000586262.1
ENST00000217515.6 |
TXNL1
|
thioredoxin-like 1 |
| chr21_-_40720995 | 0.18 |
ENST00000380749.5
|
HMGN1
|
high mobility group nucleosome binding domain 1 |
| chr17_-_7082861 | 0.18 |
ENST00000269299.3
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr4_-_141348763 | 0.17 |
ENST00000509477.1
|
CLGN
|
calmegin |
| chr16_-_28481868 | 0.17 |
ENST00000452313.1
|
NPIPB7
|
nuclear pore complex interacting protein family, member B7 |
| chr12_+_93964158 | 0.16 |
ENST00000549206.1
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chr12_-_76478446 | 0.16 |
ENST00000393263.3
ENST00000548044.1 ENST00000547704.1 ENST00000431879.3 ENST00000549596.1 ENST00000550934.1 ENST00000551600.1 ENST00000547479.1 ENST00000547773.1 ENST00000544816.1 ENST00000542344.1 ENST00000548273.1 |
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr3_+_197476621 | 0.16 |
ENST00000241502.4
|
FYTTD1
|
forty-two-three domain containing 1 |
| chr17_+_42733730 | 0.16 |
ENST00000359945.3
ENST00000425535.1 |
C17orf104
|
chromosome 17 open reading frame 104 |
| chr17_-_5322786 | 0.16 |
ENST00000225696.4
|
NUP88
|
nucleoporin 88kDa |
| chr4_+_79567057 | 0.16 |
ENST00000503259.1
ENST00000507802.1 |
RP11-792D21.2
|
long intergenic non-protein coding RNA 1094 |
| chr18_-_72265035 | 0.16 |
ENST00000585279.1
ENST00000580048.1 |
LINC00909
|
long intergenic non-protein coding RNA 909 |
| chr5_+_78407602 | 0.16 |
ENST00000274353.5
ENST00000524080.1 |
BHMT
|
betaine--homocysteine S-methyltransferase |
| chr5_-_149669192 | 0.16 |
ENST00000398376.3
|
CAMK2A
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr3_+_197477038 | 0.16 |
ENST00000426031.1
ENST00000424384.2 |
FYTTD1
|
forty-two-three domain containing 1 |
| chr7_-_37488547 | 0.16 |
ENST00000453399.1
|
ELMO1
|
engulfment and cell motility 1 |
| chr2_-_61765732 | 0.16 |
ENST00000443240.1
ENST00000436018.1 |
XPO1
|
exportin 1 (CRM1 homolog, yeast) |
| chr3_-_113464906 | 0.16 |
ENST00000477813.1
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr7_+_96745902 | 0.16 |
ENST00000432641.2
|
ACN9
|
ACN9 homolog (S. cerevisiae) |
| chr19_-_51875894 | 0.16 |
ENST00000600427.1
ENST00000595217.1 ENST00000221978.5 |
NKG7
|
natural killer cell group 7 sequence |
| chr10_+_88718314 | 0.16 |
ENST00000348795.4
|
SNCG
|
synuclein, gamma (breast cancer-specific protein 1) |
| chrX_-_119693745 | 0.15 |
ENST00000371323.2
|
CUL4B
|
cullin 4B |
| chr11_-_85430088 | 0.15 |
ENST00000533057.1
ENST00000533892.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr1_+_100817262 | 0.15 |
ENST00000455467.1
|
CDC14A
|
cell division cycle 14A |
| chr4_-_83933999 | 0.15 |
ENST00000510557.1
|
LIN54
|
lin-54 homolog (C. elegans) |
| chr12_-_122750957 | 0.15 |
ENST00000451053.2
|
VPS33A
|
vacuolar protein sorting 33 homolog A (S. cerevisiae) |
| chr16_+_56642489 | 0.15 |
ENST00000561491.1
|
MT2A
|
metallothionein 2A |
| chr15_-_65903512 | 0.15 |
ENST00000567923.1
|
VWA9
|
von Willebrand factor A domain containing 9 |
| chr1_-_54304212 | 0.15 |
ENST00000540001.1
|
NDC1
|
NDC1 transmembrane nucleoporin |
| chr8_+_120428546 | 0.14 |
ENST00000259526.3
|
NOV
|
nephroblastoma overexpressed |
| chr11_+_65265141 | 0.14 |
ENST00000534336.1
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr12_+_49687425 | 0.14 |
ENST00000257860.4
|
PRPH
|
peripherin |
| chr3_+_32726620 | 0.14 |
ENST00000331889.6
ENST00000328834.5 |
CNOT10
|
CCR4-NOT transcription complex, subunit 10 |
| chr2_-_56150184 | 0.14 |
ENST00000394554.1
|
EFEMP1
|
EGF containing fibulin-like extracellular matrix protein 1 |
| chr1_+_26348259 | 0.14 |
ENST00000374280.3
|
EXTL1
|
exostosin-like glycosyltransferase 1 |
| chr18_+_56532100 | 0.14 |
ENST00000588456.1
ENST00000589481.1 ENST00000591049.1 |
ZNF532
|
zinc finger protein 532 |
| chr19_+_50194821 | 0.14 |
ENST00000594587.1
ENST00000595969.1 |
CPT1C
|
carnitine palmitoyltransferase 1C |
| chr2_+_108905095 | 0.14 |
ENST00000251481.6
ENST00000326853.5 |
SULT1C2
|
sulfotransferase family, cytosolic, 1C, member 2 |
| chr2_-_55277692 | 0.14 |
ENST00000394611.2
|
RTN4
|
reticulon 4 |
| chr11_+_113185251 | 0.14 |
ENST00000529221.1
|
TTC12
|
tetratricopeptide repeat domain 12 |
| chr20_+_11873141 | 0.14 |
ENST00000422390.1
|
BTBD3
|
BTB (POZ) domain containing 3 |
| chr11_-_82708519 | 0.14 |
ENST00000534301.1
|
RAB30
|
RAB30, member RAS oncogene family |
| chr5_+_133562095 | 0.13 |
ENST00000602919.1
|
CTD-2410N18.3
|
CTD-2410N18.3 |
| chr19_-_51875523 | 0.13 |
ENST00000593572.1
ENST00000595157.1 |
NKG7
|
natural killer cell group 7 sequence |
| chr9_+_125703282 | 0.13 |
ENST00000373647.4
ENST00000402311.1 |
RABGAP1
|
RAB GTPase activating protein 1 |
| chr18_-_72264805 | 0.13 |
ENST00000577806.1
|
LINC00909
|
long intergenic non-protein coding RNA 909 |
| chr8_-_95908902 | 0.13 |
ENST00000520509.1
|
CCNE2
|
cyclin E2 |
| chr19_+_1105110 | 0.13 |
ENST00000587648.1
|
GPX4
|
glutathione peroxidase 4 |
| chr17_-_5015129 | 0.13 |
ENST00000575898.1
ENST00000416429.2 |
ZNF232
|
zinc finger protein 232 |
| chr20_-_43743790 | 0.13 |
ENST00000307971.4
ENST00000372789.4 |
WFDC5
|
WAP four-disulfide core domain 5 |
| chr6_+_142623063 | 0.13 |
ENST00000296932.8
ENST00000367609.3 |
GPR126
|
G protein-coupled receptor 126 |
| chr2_-_10952832 | 0.13 |
ENST00000540494.1
|
PDIA6
|
protein disulfide isomerase family A, member 6 |
| chr2_+_172290707 | 0.13 |
ENST00000375255.3
ENST00000539783.1 |
DCAF17
|
DDB1 and CUL4 associated factor 17 |
| chr2_+_198318147 | 0.13 |
ENST00000263960.2
|
COQ10B
|
coenzyme Q10 homolog B (S. cerevisiae) |
| chrX_-_106146547 | 0.12 |
ENST00000276173.4
ENST00000411805.1 |
RIPPLY1
|
ripply transcriptional repressor 1 |
| chr2_-_113542063 | 0.12 |
ENST00000263339.3
|
IL1A
|
interleukin 1, alpha |
| chr16_-_28222797 | 0.12 |
ENST00000569951.1
ENST00000565698.1 |
XPO6
|
exportin 6 |
| chr17_+_57297807 | 0.12 |
ENST00000284116.4
ENST00000581140.1 ENST00000581276.1 |
GDPD1
|
glycerophosphodiester phosphodiesterase domain containing 1 |
| chr18_+_56338618 | 0.12 |
ENST00000348428.3
|
MALT1
|
mucosa associated lymphoid tissue lymphoma translocation gene 1 |
| chr16_+_32264645 | 0.12 |
ENST00000569631.1
ENST00000354614.3 |
TP53TG3D
|
TP53 target 3D |
| chr10_-_128994422 | 0.12 |
ENST00000522781.1
|
FAM196A
|
family with sequence similarity 196, member A |
| chr7_-_151433342 | 0.12 |
ENST00000433631.2
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr7_-_151433393 | 0.12 |
ENST00000492843.1
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr6_+_155470243 | 0.12 |
ENST00000456877.2
ENST00000528391.2 |
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr6_+_3000057 | 0.12 |
ENST00000397717.2
|
NQO2
|
NAD(P)H dehydrogenase, quinone 2 |
| chr14_+_32547434 | 0.12 |
ENST00000556191.1
ENST00000554090.1 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr4_-_157892055 | 0.11 |
ENST00000422544.2
|
PDGFC
|
platelet derived growth factor C |
| chr10_+_116697946 | 0.11 |
ENST00000298746.3
|
TRUB1
|
TruB pseudouridine (psi) synthase family member 1 |
| chr11_-_92930556 | 0.11 |
ENST00000529184.1
|
SLC36A4
|
solute carrier family 36 (proton/amino acid symporter), member 4 |
| chr2_-_10952922 | 0.11 |
ENST00000272227.3
|
PDIA6
|
protein disulfide isomerase family A, member 6 |
| chr11_-_92931098 | 0.11 |
ENST00000326402.4
|
SLC36A4
|
solute carrier family 36 (proton/amino acid symporter), member 4 |
| chr7_+_76091775 | 0.11 |
ENST00000442516.1
|
DTX2
|
deltex homolog 2 (Drosophila) |
| chr17_-_40169659 | 0.11 |
ENST00000457167.4
|
DNAJC7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr4_-_80994619 | 0.11 |
ENST00000404191.1
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr11_+_46403303 | 0.11 |
ENST00000407067.1
ENST00000395565.1 |
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr2_+_108905325 | 0.11 |
ENST00000438339.1
ENST00000409880.1 ENST00000437390.2 |
SULT1C2
|
sulfotransferase family, cytosolic, 1C, member 2 |
| chrX_+_133507389 | 0.11 |
ENST00000370800.4
|
PHF6
|
PHD finger protein 6 |
| chr8_-_80680078 | 0.11 |
ENST00000337919.5
ENST00000354724.3 |
HEY1
|
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr3_+_107241882 | 0.11 |
ENST00000416476.2
|
BBX
|
bobby sox homolog (Drosophila) |
| chr12_+_41086297 | 0.11 |
ENST00000551295.2
|
CNTN1
|
contactin 1 |
| chr11_+_96123158 | 0.11 |
ENST00000332349.4
ENST00000458427.1 |
JRKL
|
jerky homolog-like (mouse) |
| chr1_-_85040090 | 0.11 |
ENST00000370630.5
|
CTBS
|
chitobiase, di-N-acetyl- |
| chr2_+_98330009 | 0.11 |
ENST00000264972.5
|
ZAP70
|
zeta-chain (TCR) associated protein kinase 70kDa |
| chr17_-_7082668 | 0.11 |
ENST00000573083.1
ENST00000574388.1 |
ASGR1
|
asialoglycoprotein receptor 1 |
| chr18_-_61089611 | 0.11 |
ENST00000591519.1
|
VPS4B
|
vacuolar protein sorting 4 homolog B (S. cerevisiae) |
| chr7_+_125078119 | 0.11 |
ENST00000458437.1
ENST00000415896.1 |
RP11-807H17.1
|
RP11-807H17.1 |
| chr22_-_37571089 | 0.10 |
ENST00000453962.1
ENST00000429622.1 ENST00000445595.1 |
IL2RB
|
interleukin 2 receptor, beta |
| chr6_-_84937314 | 0.10 |
ENST00000257766.4
ENST00000403245.3 |
KIAA1009
|
KIAA1009 |
| chr16_+_29049923 | 0.10 |
ENST00000424293.3
|
CTB-134H23.2
|
Uncharacterized protein |
| chr2_-_61765315 | 0.10 |
ENST00000406957.1
ENST00000401558.2 |
XPO1
|
exportin 1 (CRM1 homolog, yeast) |
| chr8_-_95487331 | 0.10 |
ENST00000336148.5
|
RAD54B
|
RAD54 homolog B (S. cerevisiae) |
| chr7_+_127233689 | 0.10 |
ENST00000265825.5
ENST00000420086.2 |
FSCN3
|
fascin homolog 3, actin-bundling protein, testicular (Strongylocentrotus purpuratus) |
| chr18_+_56338750 | 0.10 |
ENST00000345724.3
|
MALT1
|
mucosa associated lymphoid tissue lymphoma translocation gene 1 |
| chr11_-_85430163 | 0.10 |
ENST00000529581.1
ENST00000533577.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr10_+_19777984 | 0.10 |
ENST00000377265.3
ENST00000455457.2 |
C10orf112
|
MAM and LDL receptor class A domain containing 1 |
| chr14_+_20811722 | 0.10 |
ENST00000429687.3
|
PARP2
|
poly (ADP-ribose) polymerase 2 |
| chr3_+_50192833 | 0.10 |
ENST00000426511.1
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr22_-_32022280 | 0.10 |
ENST00000442379.1
|
PISD
|
phosphatidylserine decarboxylase |
| chr1_-_67600639 | 0.10 |
ENST00000544837.1
ENST00000603691.1 |
C1orf141
|
chromosome 1 open reading frame 141 |
| chr4_-_170533723 | 0.10 |
ENST00000510533.1
ENST00000439128.2 ENST00000511633.1 ENST00000512193.1 ENST00000507142.1 |
NEK1
|
NIMA-related kinase 1 |
| chr5_+_61708582 | 0.10 |
ENST00000325324.6
|
IPO11
|
importin 11 |
| chr2_+_198318217 | 0.10 |
ENST00000409398.1
|
COQ10B
|
coenzyme Q10 homolog B (S. cerevisiae) |
| chr1_-_11118896 | 0.10 |
ENST00000465788.1
|
SRM
|
spermidine synthase |
| chr4_-_141348789 | 0.10 |
ENST00000414773.1
|
CLGN
|
calmegin |
| chr1_-_236046872 | 0.10 |
ENST00000536965.1
|
LYST
|
lysosomal trafficking regulator |
| chr7_-_105162652 | 0.09 |
ENST00000356362.2
ENST00000469408.1 |
PUS7
|
pseudouridylate synthase 7 homolog (S. cerevisiae) |
| chr14_+_20811766 | 0.09 |
ENST00000250416.5
ENST00000527915.1 |
PARP2
|
poly (ADP-ribose) polymerase 2 |
| chr11_-_93583697 | 0.09 |
ENST00000409977.1
|
VSTM5
|
V-set and transmembrane domain containing 5 |
| chr5_+_54455946 | 0.09 |
ENST00000503787.1
ENST00000296734.6 ENST00000515370.1 |
GPX8
|
glutathione peroxidase 8 (putative) |
| chr3_-_119396193 | 0.09 |
ENST00000484810.1
ENST00000497116.1 ENST00000261070.2 |
COX17
|
COX17 cytochrome c oxidase copper chaperone |
| chr19_-_4338838 | 0.09 |
ENST00000594605.1
|
STAP2
|
signal transducing adaptor family member 2 |
| chr19_+_38826415 | 0.09 |
ENST00000410018.1
ENST00000409235.3 |
CATSPERG
|
catsper channel auxiliary subunit gamma |
| chr1_+_35247859 | 0.09 |
ENST00000373362.3
|
GJB3
|
gap junction protein, beta 3, 31kDa |
| chr10_+_6625733 | 0.09 |
ENST00000607982.1
ENST00000608526.1 |
PRKCQ-AS1
|
PRKCQ antisense RNA 1 |
| chr5_-_111093406 | 0.09 |
ENST00000379671.3
|
NREP
|
neuronal regeneration related protein |
| chr11_+_65554493 | 0.09 |
ENST00000335987.3
|
OVOL1
|
ovo-like zinc finger 1 |
| chr6_-_8102714 | 0.09 |
ENST00000502429.1
ENST00000429723.2 ENST00000507463.1 ENST00000379715.5 |
EEF1E1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr3_-_142297668 | 0.09 |
ENST00000350721.4
ENST00000383101.3 |
ATR
|
ataxia telangiectasia and Rad3 related |
| chr3_-_42845951 | 0.09 |
ENST00000418900.2
ENST00000430190.1 |
HIGD1A
|
HIG1 hypoxia inducible domain family, member 1A |
| chr16_-_82045049 | 0.09 |
ENST00000532128.1
ENST00000328945.5 |
SDR42E1
|
short chain dehydrogenase/reductase family 42E, member 1 |
| chr2_-_55277436 | 0.09 |
ENST00000354474.6
|
RTN4
|
reticulon 4 |
| chr22_+_45898712 | 0.09 |
ENST00000455233.1
ENST00000348697.2 ENST00000402984.3 ENST00000262722.7 ENST00000327858.6 ENST00000442170.2 ENST00000340923.5 ENST00000439835.1 |
FBLN1
|
fibulin 1 |
| chr4_-_99850243 | 0.09 |
ENST00000280892.6
ENST00000511644.1 ENST00000504432.1 ENST00000505992.1 |
EIF4E
|
eukaryotic translation initiation factor 4E |
| chrX_+_133507327 | 0.09 |
ENST00000332070.3
ENST00000394292.1 ENST00000370799.1 ENST00000416404.2 |
PHF6
|
PHD finger protein 6 |
| chr14_-_24685246 | 0.09 |
ENST00000396833.2
ENST00000288087.7 |
MDP1
|
magnesium-dependent phosphatase 1 |
| chr7_-_6048702 | 0.09 |
ENST00000265849.7
|
PMS2
|
PMS2 postmeiotic segregation increased 2 (S. cerevisiae) |
| chr6_+_30978251 | 0.09 |
ENST00000561890.1
|
MUC22
|
mucin 22 |
| chr15_-_34635314 | 0.09 |
ENST00000557912.1
ENST00000328848.4 |
NOP10
|
NOP10 ribonucleoprotein |
| chr19_+_8455077 | 0.09 |
ENST00000328024.6
|
RAB11B
|
RAB11B, member RAS oncogene family |
| chr2_+_200820269 | 0.09 |
ENST00000392290.1
|
C2orf47
|
chromosome 2 open reading frame 47 |
| chr12_+_27397166 | 0.09 |
ENST00000545470.1
ENST00000540996.1 ENST00000539577.1 |
STK38L
|
serine/threonine kinase 38 like |
| chr2_+_75874105 | 0.08 |
ENST00000476622.1
|
MRPL19
|
mitochondrial ribosomal protein L19 |
| chr5_+_34757309 | 0.08 |
ENST00000397449.1
|
RAI14
|
retinoic acid induced 14 |
| chr14_-_70826438 | 0.08 |
ENST00000389912.6
|
COX16
|
COX16 cytochrome c oxidase assembly homolog (S. cerevisiae) |
| chr7_+_141490017 | 0.08 |
ENST00000247883.4
|
TAS2R5
|
taste receptor, type 2, member 5 |
| chr9_-_130213652 | 0.08 |
ENST00000536368.1
ENST00000361436.5 |
RPL12
|
ribosomal protein L12 |
| chr16_+_86609939 | 0.08 |
ENST00000593625.1
|
FOXL1
|
forkhead box L1 |
| chr22_+_32754139 | 0.08 |
ENST00000382088.3
|
RFPL3
|
ret finger protein-like 3 |
| chr5_+_137225125 | 0.08 |
ENST00000350250.4
ENST00000508638.1 ENST00000502810.1 ENST00000508883.1 |
PKD2L2
|
polycystic kidney disease 2-like 2 |
| chr11_-_122931881 | 0.08 |
ENST00000526110.1
ENST00000227378.3 |
HSPA8
|
heat shock 70kDa protein 8 |
| chr18_-_55470320 | 0.08 |
ENST00000536015.1
|
ATP8B1
|
ATPase, aminophospholipid transporter, class I, type 8B, member 1 |
| chr15_+_65903680 | 0.08 |
ENST00000537259.1
|
SLC24A1
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 1 |
| chr12_-_32040123 | 0.08 |
ENST00000535163.1
|
RP11-428G5.5
|
RP11-428G5.5 |
| chr2_+_103089756 | 0.08 |
ENST00000295269.4
|
SLC9A4
|
solute carrier family 9, subfamily A (NHE4, cation proton antiporter 4), member 4 |
| chr6_+_31371337 | 0.08 |
ENST00000449934.2
ENST00000421350.1 |
MICA
|
MHC class I polypeptide-related sequence A |
| chr1_-_27709793 | 0.08 |
ENST00000374027.3
ENST00000374025.3 |
CD164L2
|
CD164 sialomucin-like 2 |
| chr5_-_39219705 | 0.08 |
ENST00000351578.6
|
FYB
|
FYN binding protein |
| chr11_+_114271314 | 0.08 |
ENST00000541475.1
|
RBM7
|
RNA binding motif protein 7 |
| chr7_-_120497178 | 0.08 |
ENST00000441017.1
ENST00000424710.1 ENST00000433758.1 |
TSPAN12
|
tetraspanin 12 |
| chr3_+_108321623 | 0.08 |
ENST00000497905.1
ENST00000463306.1 |
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr9_-_104198042 | 0.08 |
ENST00000374855.4
|
ALDOB
|
aldolase B, fructose-bisphosphate |
| chr4_-_129207942 | 0.08 |
ENST00000503588.1
|
PGRMC2
|
progesterone receptor membrane component 2 |
| chr16_+_4666475 | 0.08 |
ENST00000591895.1
|
MGRN1
|
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr1_-_212588157 | 0.08 |
ENST00000261455.4
ENST00000535273.1 |
TMEM206
|
transmembrane protein 206 |
| chr5_-_139682658 | 0.07 |
ENST00000524074.1
ENST00000510217.1 ENST00000261813.4 |
PFDN1
|
prefoldin subunit 1 |
| chr14_-_71107921 | 0.07 |
ENST00000553982.1
ENST00000500016.1 |
CTD-2540L5.5
CTD-2540L5.6
|
CTD-2540L5.5 CTD-2540L5.6 |
| chr11_+_114271251 | 0.07 |
ENST00000375490.5
|
RBM7
|
RNA binding motif protein 7 |
| chr6_+_107349392 | 0.07 |
ENST00000443043.1
ENST00000405204.2 ENST00000311381.5 |
C6orf203
|
chromosome 6 open reading frame 203 |
| chr1_-_21503337 | 0.07 |
ENST00000400422.1
ENST00000602326.1 ENST00000411888.1 ENST00000438975.1 |
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr10_-_128359074 | 0.07 |
ENST00000544758.1
|
C10orf90
|
chromosome 10 open reading frame 90 |
| chr6_+_37012607 | 0.07 |
ENST00000423336.1
|
COX6A1P2
|
cytochrome c oxidase subunit VIa polypeptide 1 pseudogene 2 |
| chr11_+_114270752 | 0.07 |
ENST00000540163.1
|
RBM7
|
RNA binding motif protein 7 |
| chr13_-_114843416 | 0.07 |
ENST00000389544.4
|
RASA3
|
RAS p21 protein activator 3 |
| chr14_-_68157084 | 0.07 |
ENST00000557564.1
|
RP11-1012A1.4
|
RP11-1012A1.4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TFCP2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 0.0 | 0.6 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.2 | GO:0035188 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.2 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.0 | 0.1 | GO:0030474 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.0 | 0.2 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.0 | 0.4 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.2 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.0 | 0.1 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 | 0.1 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.2 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.0 | 0.1 | GO:1901994 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.1 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.0 | 0.1 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.0 | 0.2 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.1 | GO:1903722 | negative regulation of exosomal secretion(GO:1903542) regulation of centriole elongation(GO:1903722) |
| 0.0 | 0.2 | GO:0035524 | L-alanine transport(GO:0015808) proline transmembrane transport(GO:0035524) |
| 0.0 | 0.3 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.1 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.1 | GO:2001193 | gamma-delta T cell activation involved in immune response(GO:0002290) negative regulation of interferon-beta secretion(GO:0035548) regulation of gamma-delta T cell activation involved in immune response(GO:2001191) positive regulation of gamma-delta T cell activation involved in immune response(GO:2001193) |
| 0.0 | 0.2 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.2 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.5 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.0 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.1 | GO:1901656 | cellular response to mycotoxin(GO:0036146) glycoside transport(GO:1901656) cellular response to bile acid(GO:1903413) |
| 0.0 | 0.1 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.0 | 0.1 | GO:1904882 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.0 | 0.1 | GO:0043366 | beta selection(GO:0043366) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.3 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.1 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.1 | GO:1904764 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.0 | 0.0 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.1 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.0 | 0.1 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.1 | GO:0007260 | tyrosine phosphorylation of STAT protein(GO:0007260) regulation of tyrosine phosphorylation of STAT protein(GO:0042509) positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) positive regulation of JAK-STAT cascade(GO:0046427) positive regulation of STAT cascade(GO:1904894) |
| 0.0 | 0.1 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 0.1 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.1 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.0 | 0.2 | GO:0035290 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.0 | 0.0 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.0 | 0.1 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.0 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.0 | 0.4 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 0.1 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.0 | 0.2 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.0 | 0.2 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.1 | GO:0019867 | outer membrane(GO:0019867) |
| 0.0 | 0.1 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.0 | 0.2 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.1 | GO:0005882 | intermediate filament(GO:0005882) intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 0.1 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.1 | GO:0090661 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.1 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.2 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.1 | 0.5 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.1 | 0.2 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.1 | 0.3 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.2 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.2 | GO:1904408 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 0.0 | 0.2 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.1 | GO:0052812 | phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.1 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.0 | 0.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.0 | 0.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.0 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.1 | GO:0030107 | HLA-A specific inhibitory MHC class I receptor activity(GO:0030107) HLA-B specific inhibitory MHC class I receptor activity(GO:0030109) inhibitory MHC class I receptor activity(GO:0032396) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.2 | GO:0032407 | MutSalpha complex binding(GO:0032407) |
| 0.0 | 0.1 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 0.0 | 0.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.1 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.0 | 0.2 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.1 | GO:0015207 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.1 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.6 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |