Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
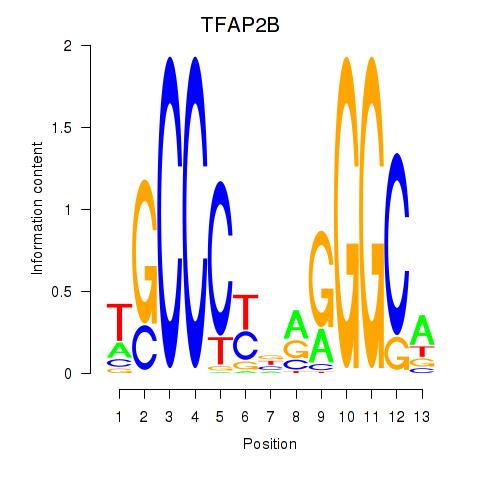
Results for TFAP2B
Z-value: 0.63
Transcription factors associated with TFAP2B
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TFAP2B
|
ENSG00000008196.8 | transcription factor AP-2 beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TFAP2B | hg19_v2_chr6_+_50786414_50786439 | -0.95 | 5.5e-02 | Click! |
Activity profile of TFAP2B motif
Sorted Z-values of TFAP2B motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_100065686 | 0.65 |
ENST00000423266.1
ENST00000456330.1 |
TSC22D4
|
TSC22 domain family, member 4 |
| chr4_-_1723040 | 0.64 |
ENST00000382936.3
ENST00000536901.1 ENST00000303277.2 |
TMEM129
|
transmembrane protein 129 |
| chr6_+_33378517 | 0.51 |
ENST00000428274.1
|
PHF1
|
PHD finger protein 1 |
| chr9_+_130922537 | 0.48 |
ENST00000372994.1
|
C9orf16
|
chromosome 9 open reading frame 16 |
| chr2_-_220408260 | 0.46 |
ENST00000373891.2
|
CHPF
|
chondroitin polymerizing factor |
| chr11_-_64052111 | 0.44 |
ENST00000394532.3
ENST00000394531.3 ENST00000309032.3 |
BAD
|
BCL2-associated agonist of cell death |
| chr17_-_8093471 | 0.43 |
ENST00000389017.4
|
C17orf59
|
chromosome 17 open reading frame 59 |
| chr19_+_41107249 | 0.40 |
ENST00000396819.3
|
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chr2_-_220408430 | 0.39 |
ENST00000243776.6
|
CHPF
|
chondroitin polymerizing factor |
| chr19_+_47778119 | 0.36 |
ENST00000552360.2
|
PRR24
|
proline rich 24 |
| chr8_-_21988558 | 0.34 |
ENST00000312841.8
|
HR
|
hair growth associated |
| chr10_+_104178946 | 0.32 |
ENST00000432590.1
|
FBXL15
|
F-box and leucine-rich repeat protein 15 |
| chr16_-_28223229 | 0.31 |
ENST00000566073.1
|
XPO6
|
exportin 6 |
| chr6_+_33378738 | 0.30 |
ENST00000374512.3
ENST00000374516.3 |
PHF1
|
PHD finger protein 1 |
| chr17_-_61777459 | 0.30 |
ENST00000578993.1
ENST00000583211.1 ENST00000259006.3 |
LIMD2
|
LIM domain containing 2 |
| chr1_+_155179012 | 0.29 |
ENST00000609421.1
|
MTX1
|
metaxin 1 |
| chr8_+_21906433 | 0.29 |
ENST00000522148.1
|
DMTN
|
dematin actin binding protein |
| chr17_+_18163848 | 0.28 |
ENST00000323019.4
ENST00000578174.1 ENST00000395704.4 ENST00000395703.4 ENST00000578621.1 ENST00000579341.1 |
MIEF2
|
mitochondrial elongation factor 2 |
| chr6_-_31763276 | 0.27 |
ENST00000440048.1
|
VARS
|
valyl-tRNA synthetase |
| chr1_-_1677358 | 0.27 |
ENST00000355439.2
ENST00000400924.1 ENST00000246421.4 |
SLC35E2
|
solute carrier family 35, member E2 |
| chr5_-_176730733 | 0.26 |
ENST00000504395.1
|
RAB24
|
RAB24, member RAS oncogene family |
| chr17_+_1627834 | 0.26 |
ENST00000419248.1
ENST00000418841.1 |
WDR81
|
WD repeat domain 81 |
| chr2_+_130939827 | 0.26 |
ENST00000409255.1
ENST00000455239.1 |
MZT2B
|
mitotic spindle organizing protein 2B |
| chr1_+_155178481 | 0.26 |
ENST00000368376.3
|
MTX1
|
metaxin 1 |
| chr1_-_2345236 | 0.25 |
ENST00000508384.1
|
PEX10
|
peroxisomal biogenesis factor 10 |
| chr17_-_17726907 | 0.25 |
ENST00000423161.3
|
SREBF1
|
sterol regulatory element binding transcription factor 1 |
| chr19_+_7710774 | 0.24 |
ENST00000602355.1
|
STXBP2
|
syntaxin binding protein 2 |
| chr2_+_232575168 | 0.23 |
ENST00000440384.1
|
PTMA
|
prothymosin, alpha |
| chr17_-_7760457 | 0.23 |
ENST00000576384.1
|
LSMD1
|
LSM domain containing 1 |
| chr19_+_13228917 | 0.23 |
ENST00000586171.1
|
NACC1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr12_+_48152774 | 0.22 |
ENST00000549243.1
|
SLC48A1
|
solute carrier family 48 (heme transporter), member 1 |
| chr5_+_172068232 | 0.22 |
ENST00000520919.1
ENST00000522853.1 ENST00000369800.5 |
NEURL1B
|
neuralized E3 ubiquitin protein ligase 1B |
| chr1_+_155178518 | 0.22 |
ENST00000316721.4
|
MTX1
|
metaxin 1 |
| chr8_+_142402089 | 0.22 |
ENST00000521578.1
ENST00000520105.1 ENST00000523147.1 |
PTP4A3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr20_+_62327996 | 0.22 |
ENST00000369996.1
|
TNFRSF6B
|
tumor necrosis factor receptor superfamily, member 6b, decoy |
| chr14_+_24458093 | 0.22 |
ENST00000558753.1
ENST00000537912.1 |
DHRS4L2
|
dehydrogenase/reductase (SDR family) member 4 like 2 |
| chr7_-_86849836 | 0.21 |
ENST00000455575.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr19_-_46145696 | 0.21 |
ENST00000588172.1
|
EML2
|
echinoderm microtubule associated protein like 2 |
| chr16_+_30960375 | 0.21 |
ENST00000318663.4
ENST00000566237.1 ENST00000562699.1 |
ORAI3
|
ORAI calcium release-activated calcium modulator 3 |
| chr10_+_103825080 | 0.21 |
ENST00000299238.5
|
HPS6
|
Hermansky-Pudlak syndrome 6 |
| chr17_-_72869086 | 0.21 |
ENST00000581530.1
ENST00000420580.2 ENST00000455107.2 ENST00000413947.2 ENST00000581219.1 ENST00000582944.1 |
FDXR
|
ferredoxin reductase |
| chr17_-_72869140 | 0.20 |
ENST00000583917.1
ENST00000293195.5 ENST00000442102.2 |
FDXR
|
ferredoxin reductase |
| chr20_-_49639631 | 0.20 |
ENST00000424171.1
ENST00000439216.1 ENST00000371571.4 |
KCNG1
|
potassium voltage-gated channel, subfamily G, member 1 |
| chr14_+_65171099 | 0.20 |
ENST00000247226.7
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr9_+_135037334 | 0.19 |
ENST00000393229.3
ENST00000360670.3 ENST00000393228.4 ENST00000372179.3 |
NTNG2
|
netrin G2 |
| chr11_+_62379194 | 0.19 |
ENST00000525801.1
ENST00000534093.1 |
ROM1
|
retinal outer segment membrane protein 1 |
| chr1_-_44497118 | 0.19 |
ENST00000537678.1
ENST00000466926.1 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr22_+_42665742 | 0.19 |
ENST00000332965.3
ENST00000415205.1 ENST00000446578.1 |
Z83851.3
|
Z83851.3 |
| chr20_-_30539773 | 0.19 |
ENST00000202017.4
|
PDRG1
|
p53 and DNA-damage regulated 1 |
| chr9_+_136325089 | 0.18 |
ENST00000291722.7
ENST00000316948.4 ENST00000540581.1 |
CACFD1
|
calcium channel flower domain containing 1 |
| chr6_+_30294186 | 0.18 |
ENST00000458516.1
|
TRIM39
|
tripartite motif containing 39 |
| chr19_+_39989535 | 0.18 |
ENST00000356433.5
|
DLL3
|
delta-like 3 (Drosophila) |
| chr5_-_134369973 | 0.18 |
ENST00000265340.7
|
PITX1
|
paired-like homeodomain 1 |
| chr1_-_21948906 | 0.18 |
ENST00000374761.2
ENST00000599760.1 |
RAP1GAP
|
RAP1 GTPase activating protein |
| chr1_-_156399184 | 0.17 |
ENST00000368243.1
ENST00000357975.4 ENST00000310027.5 ENST00000400991.2 |
C1orf61
|
chromosome 1 open reading frame 61 |
| chr1_-_1535455 | 0.17 |
ENST00000422725.1
|
C1orf233
|
chromosome 1 open reading frame 233 |
| chr2_-_132249955 | 0.17 |
ENST00000309451.6
|
MZT2A
|
mitotic spindle organizing protein 2A |
| chr11_+_809647 | 0.17 |
ENST00000321153.4
|
RPLP2
|
ribosomal protein, large, P2 |
| chr16_+_88772866 | 0.17 |
ENST00000453996.2
ENST00000312060.5 ENST00000378384.3 ENST00000567949.1 ENST00000564921.1 |
CTU2
|
cytosolic thiouridylase subunit 2 homolog (S. pombe) |
| chr2_-_73511559 | 0.17 |
ENST00000521871.1
|
FBXO41
|
F-box protein 41 |
| chr19_+_14544099 | 0.17 |
ENST00000242783.6
ENST00000586557.1 ENST00000590097.1 |
PKN1
|
protein kinase N1 |
| chr22_+_45072925 | 0.17 |
ENST00000006251.7
|
PRR5
|
proline rich 5 (renal) |
| chr17_-_73874654 | 0.17 |
ENST00000254816.2
|
TRIM47
|
tripartite motif containing 47 |
| chr14_+_23341513 | 0.17 |
ENST00000546834.1
|
LRP10
|
low density lipoprotein receptor-related protein 10 |
| chr11_-_62379752 | 0.16 |
ENST00000466671.1
ENST00000466886.1 |
EML3
|
echinoderm microtubule associated protein like 3 |
| chr3_-_50360192 | 0.16 |
ENST00000442581.1
ENST00000447092.1 ENST00000357750.4 |
HYAL2
|
hyaluronoglucosaminidase 2 |
| chr19_+_13229126 | 0.16 |
ENST00000292431.4
|
NACC1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr19_+_41119794 | 0.16 |
ENST00000593463.1
|
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chr17_+_36861735 | 0.16 |
ENST00000378137.5
ENST00000325718.7 |
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6 |
| chr15_+_73976715 | 0.16 |
ENST00000558689.1
ENST00000560786.2 ENST00000561213.1 ENST00000563584.1 ENST00000561416.1 |
CD276
|
CD276 molecule |
| chr8_-_145018905 | 0.16 |
ENST00000398774.2
|
PLEC
|
plectin |
| chr17_-_7760779 | 0.16 |
ENST00000335155.5
ENST00000575071.1 |
LSMD1
|
LSM domain containing 1 |
| chr22_-_19842330 | 0.16 |
ENST00000416337.1
ENST00000328554.4 ENST00000403325.1 ENST00000453108.1 |
C22orf29
GNB1L
|
chromosome 22 open reading frame 29 guanine nucleotide binding protein (G protein), beta polypeptide 1-like |
| chr16_-_67970990 | 0.16 |
ENST00000358514.4
|
PSMB10
|
proteasome (prosome, macropain) subunit, beta type, 10 |
| chrX_+_48916497 | 0.16 |
ENST00000496529.2
ENST00000376396.3 ENST00000422185.2 ENST00000603986.1 ENST00000536628.2 |
CCDC120
|
coiled-coil domain containing 120 |
| chr19_-_42724261 | 0.16 |
ENST00000595337.1
|
DEDD2
|
death effector domain containing 2 |
| chr12_+_133067157 | 0.16 |
ENST00000261673.6
|
FBRSL1
|
fibrosin-like 1 |
| chr17_-_71308119 | 0.16 |
ENST00000439510.2
ENST00000581014.1 ENST00000579611.1 |
CDC42EP4
|
CDC42 effector protein (Rho GTPase binding) 4 |
| chr6_-_34664612 | 0.15 |
ENST00000374023.3
ENST00000374026.3 |
C6orf106
|
chromosome 6 open reading frame 106 |
| chr22_+_45072958 | 0.15 |
ENST00000403581.1
|
PRR5
|
proline rich 5 (renal) |
| chr3_-_10052849 | 0.15 |
ENST00000437616.1
ENST00000429065.2 |
AC022007.5
|
AC022007.5 |
| chr14_+_23340822 | 0.15 |
ENST00000359591.4
|
LRP10
|
low density lipoprotein receptor-related protein 10 |
| chr19_+_46144884 | 0.15 |
ENST00000593161.1
|
AC006132.1
|
chromosome 19 open reading frame 83 |
| chr22_-_29663690 | 0.14 |
ENST00000406335.1
|
RHBDD3
|
rhomboid domain containing 3 |
| chr17_-_27230035 | 0.14 |
ENST00000378895.4
ENST00000394901.3 |
DHRS13
|
dehydrogenase/reductase (SDR family) member 13 |
| chr12_+_110011571 | 0.14 |
ENST00000539696.1
ENST00000228510.3 ENST00000392727.3 |
MVK
|
mevalonate kinase |
| chrX_+_70316005 | 0.14 |
ENST00000374259.3
|
FOXO4
|
forkhead box O4 |
| chr8_-_146017736 | 0.14 |
ENST00000528957.1
|
RPL8
|
ribosomal protein L8 |
| chr9_+_116917807 | 0.14 |
ENST00000356083.3
|
COL27A1
|
collagen, type XXVII, alpha 1 |
| chr16_+_30418910 | 0.14 |
ENST00000566625.1
|
ZNF771
|
zinc finger protein 771 |
| chr3_-_185216766 | 0.14 |
ENST00000296254.3
|
TMEM41A
|
transmembrane protein 41A |
| chr1_+_155051379 | 0.14 |
ENST00000418360.2
|
EFNA3
|
ephrin-A3 |
| chr5_-_176981417 | 0.14 |
ENST00000514747.1
ENST00000443375.2 ENST00000329540.5 |
FAM193B
|
family with sequence similarity 193, member B |
| chr11_-_63536113 | 0.14 |
ENST00000433688.1
ENST00000546282.2 |
C11orf95
RP11-466C23.4
|
chromosome 11 open reading frame 95 RP11-466C23.4 |
| chr16_+_31085714 | 0.14 |
ENST00000300850.5
ENST00000564189.1 ENST00000428260.1 |
ZNF646
|
zinc finger protein 646 |
| chr1_+_33207381 | 0.13 |
ENST00000401073.2
|
KIAA1522
|
KIAA1522 |
| chr12_+_53693466 | 0.13 |
ENST00000267103.5
ENST00000548632.1 |
C12orf10
|
chromosome 12 open reading frame 10 |
| chr16_-_88772761 | 0.13 |
ENST00000567844.1
ENST00000312838.4 |
RNF166
|
ring finger protein 166 |
| chr17_+_7255208 | 0.13 |
ENST00000333751.3
|
KCTD11
|
potassium channel tetramerization domain containing 11 |
| chr10_-_135238076 | 0.13 |
ENST00000414069.2
|
SPRN
|
shadow of prion protein homolog (zebrafish) |
| chr1_-_44497024 | 0.13 |
ENST00000372306.3
ENST00000372310.3 ENST00000475075.2 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr7_+_65939543 | 0.13 |
ENST00000600021.1
|
AC008267.1
|
HCG1983814; Uncharacterized protein |
| chr7_+_150076406 | 0.13 |
ENST00000329630.5
|
ZNF775
|
zinc finger protein 775 |
| chr9_-_131534160 | 0.13 |
ENST00000291900.2
|
ZER1
|
zyg-11 related, cell cycle regulator |
| chr6_+_43139037 | 0.13 |
ENST00000265354.4
|
SRF
|
serum response factor (c-fos serum response element-binding transcription factor) |
| chr17_+_36858694 | 0.13 |
ENST00000563897.1
|
CTB-58E17.1
|
CTB-58E17.1 |
| chr17_-_48133054 | 0.13 |
ENST00000499842.1
|
RP11-1094H24.4
|
RP11-1094H24.4 |
| chr1_+_33722080 | 0.13 |
ENST00000483388.1
ENST00000539719.1 |
ZNF362
|
zinc finger protein 362 |
| chr19_-_42758040 | 0.13 |
ENST00000593944.1
|
ERF
|
Ets2 repressor factor |
| chr9_+_136325149 | 0.13 |
ENST00000542192.1
|
CACFD1
|
calcium channel flower domain containing 1 |
| chr19_-_39832563 | 0.13 |
ENST00000599274.1
|
CTC-246B18.10
|
CTC-246B18.10 |
| chr9_+_131182697 | 0.13 |
ENST00000372838.4
ENST00000411852.1 |
CERCAM
|
cerebral endothelial cell adhesion molecule |
| chr14_+_65171315 | 0.13 |
ENST00000394691.1
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr3_-_50360165 | 0.12 |
ENST00000428028.1
|
HYAL2
|
hyaluronoglucosaminidase 2 |
| chr19_+_39989580 | 0.12 |
ENST00000596614.1
ENST00000205143.4 |
DLL3
|
delta-like 3 (Drosophila) |
| chr12_+_6493319 | 0.12 |
ENST00000536876.1
|
LTBR
|
lymphotoxin beta receptor (TNFR superfamily, member 3) |
| chr8_-_146017765 | 0.12 |
ENST00000532702.1
ENST00000394920.2 ENST00000527914.1 |
RPL8
|
ribosomal protein L8 |
| chr1_+_161129240 | 0.12 |
ENST00000492950.1
|
USP21
|
ubiquitin specific peptidase 21 |
| chr22_-_31742218 | 0.12 |
ENST00000266269.5
ENST00000405309.3 ENST00000351933.4 |
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr5_+_149980622 | 0.12 |
ENST00000394243.1
|
SYNPO
|
synaptopodin |
| chr16_+_57126428 | 0.12 |
ENST00000290776.8
|
CPNE2
|
copine II |
| chr10_+_99079008 | 0.12 |
ENST00000371021.3
|
FRAT1
|
frequently rearranged in advanced T-cell lymphomas |
| chr9_-_140009130 | 0.12 |
ENST00000497375.1
ENST00000371579.2 |
DPP7
|
dipeptidyl-peptidase 7 |
| chr16_+_3014269 | 0.12 |
ENST00000575885.1
ENST00000571007.1 ENST00000319500.6 |
KREMEN2
|
kringle containing transmembrane protein 2 |
| chr17_+_46985823 | 0.12 |
ENST00000508468.2
|
UBE2Z
|
ubiquitin-conjugating enzyme E2Z |
| chr20_+_44462749 | 0.12 |
ENST00000372541.1
|
SNX21
|
sorting nexin family member 21 |
| chr1_+_45792541 | 0.12 |
ENST00000334815.3
|
HPDL
|
4-hydroxyphenylpyruvate dioxygenase-like |
| chr9_-_131534188 | 0.12 |
ENST00000414921.1
|
ZER1
|
zyg-11 related, cell cycle regulator |
| chr12_+_6493199 | 0.12 |
ENST00000228918.4
|
LTBR
|
lymphotoxin beta receptor (TNFR superfamily, member 3) |
| chr19_-_10679644 | 0.12 |
ENST00000393599.2
|
CDKN2D
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
| chr1_+_9299895 | 0.12 |
ENST00000602477.1
|
H6PD
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr22_-_36018569 | 0.12 |
ENST00000419229.1
ENST00000406324.1 |
MB
|
myoglobin |
| chr19_+_42724423 | 0.12 |
ENST00000301215.3
ENST00000597945.1 |
ZNF526
|
zinc finger protein 526 |
| chr18_+_3450161 | 0.12 |
ENST00000551402.1
ENST00000577543.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr1_-_246670614 | 0.12 |
ENST00000403792.3
|
SMYD3
|
SET and MYND domain containing 3 |
| chr17_+_7761013 | 0.12 |
ENST00000571846.1
|
CYB5D1
|
cytochrome b5 domain containing 1 |
| chr1_-_167905225 | 0.11 |
ENST00000367846.4
|
MPC2
|
mitochondrial pyruvate carrier 2 |
| chr11_-_62380199 | 0.11 |
ENST00000419857.1
ENST00000394773.2 |
EML3
|
echinoderm microtubule associated protein like 3 |
| chrX_+_152240819 | 0.11 |
ENST00000421798.3
ENST00000535416.1 |
PNMA6C
PNMA6A
|
paraneoplastic Ma antigen family member 6C paraneoplastic Ma antigen family member 6A |
| chr19_+_39833036 | 0.11 |
ENST00000602243.1
ENST00000598913.1 ENST00000314471.6 |
SAMD4B
|
sterile alpha motif domain containing 4B |
| chr19_-_49149553 | 0.11 |
ENST00000084798.4
|
CA11
|
carbonic anhydrase XI |
| chr16_+_2285817 | 0.11 |
ENST00000564065.1
|
DNASE1L2
|
deoxyribonuclease I-like 2 |
| chr10_-_35930219 | 0.11 |
ENST00000374694.1
|
FZD8
|
frizzled family receptor 8 |
| chr14_+_65170820 | 0.11 |
ENST00000555982.1
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr14_+_24422795 | 0.11 |
ENST00000313250.5
ENST00000558263.1 ENST00000543741.2 ENST00000421831.1 ENST00000397073.2 ENST00000308178.8 ENST00000382761.3 ENST00000397075.3 ENST00000397074.3 ENST00000559632.1 ENST00000558581.1 |
DHRS4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr19_-_19739321 | 0.11 |
ENST00000588461.1
|
LPAR2
|
lysophosphatidic acid receptor 2 |
| chr20_-_49639612 | 0.11 |
ENST00000396017.3
ENST00000433903.1 |
KCNG1
|
potassium voltage-gated channel, subfamily G, member 1 |
| chr12_-_122238913 | 0.11 |
ENST00000537157.1
|
AC084018.1
|
AC084018.1 |
| chr15_+_90319557 | 0.11 |
ENST00000341735.3
|
MESP2
|
mesoderm posterior 2 homolog (mouse) |
| chr22_-_27620603 | 0.11 |
ENST00000418271.1
ENST00000444114.1 |
RP5-1172A22.1
|
RP5-1172A22.1 |
| chr6_-_33385854 | 0.11 |
ENST00000488478.1
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr17_-_2304365 | 0.11 |
ENST00000575394.1
ENST00000174618.4 |
MNT
|
MAX network transcriptional repressor |
| chr8_+_22462145 | 0.11 |
ENST00000308511.4
ENST00000523801.1 ENST00000521301.1 |
CCAR2
|
cell cycle and apoptosis regulator 2 |
| chr16_-_75285380 | 0.11 |
ENST00000393420.6
ENST00000162330.5 |
BCAR1
|
breast cancer anti-estrogen resistance 1 |
| chr11_+_47279155 | 0.11 |
ENST00000444396.1
ENST00000457932.1 ENST00000412937.1 |
NR1H3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr11_-_62379994 | 0.10 |
ENST00000278845.4
ENST00000529309.1 |
EML3
|
echinoderm microtubule associated protein like 3 |
| chr15_+_75335604 | 0.10 |
ENST00000563393.1
|
PPCDC
|
phosphopantothenoylcysteine decarboxylase |
| chr22_+_30115986 | 0.10 |
ENST00000216144.3
|
CABP7
|
calcium binding protein 7 |
| chr22_-_20255212 | 0.10 |
ENST00000416372.1
|
RTN4R
|
reticulon 4 receptor |
| chr17_-_79869340 | 0.10 |
ENST00000538936.2
|
PCYT2
|
phosphate cytidylyltransferase 2, ethanolamine |
| chr19_+_42387228 | 0.10 |
ENST00000354532.3
ENST00000599846.1 ENST00000347545.4 |
ARHGEF1
|
Rho guanine nucleotide exchange factor (GEF) 1 |
| chr3_+_44903361 | 0.10 |
ENST00000302392.4
|
TMEM42
|
transmembrane protein 42 |
| chr17_+_48638371 | 0.10 |
ENST00000360761.4
ENST00000352832.5 ENST00000354983.4 |
CACNA1G
|
calcium channel, voltage-dependent, T type, alpha 1G subunit |
| chr9_+_35605274 | 0.10 |
ENST00000336395.5
|
TESK1
|
testis-specific kinase 1 |
| chr17_-_33446820 | 0.10 |
ENST00000592577.1
ENST00000590016.1 ENST00000345365.6 ENST00000360276.3 ENST00000357906.3 |
RAD51D
|
RAD51 paralog D |
| chr13_-_99852916 | 0.10 |
ENST00000426037.2
ENST00000445737.2 |
UBAC2-AS1
|
UBAC2 antisense RNA 1 |
| chr19_+_38397839 | 0.10 |
ENST00000222345.6
|
SIPA1L3
|
signal-induced proliferation-associated 1 like 3 |
| chr2_+_26915584 | 0.10 |
ENST00000302909.3
|
KCNK3
|
potassium channel, subfamily K, member 3 |
| chr6_-_30654977 | 0.10 |
ENST00000399199.3
|
PPP1R18
|
protein phosphatase 1, regulatory subunit 18 |
| chr16_-_66586365 | 0.10 |
ENST00000562484.2
|
TK2
|
thymidine kinase 2, mitochondrial |
| chr1_+_160370344 | 0.10 |
ENST00000368061.2
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr22_-_46933067 | 0.10 |
ENST00000262738.3
ENST00000395964.1 |
CELSR1
|
cadherin, EGF LAG seven-pass G-type receptor 1 |
| chr19_-_19774473 | 0.10 |
ENST00000357324.6
|
ATP13A1
|
ATPase type 13A1 |
| chr12_-_58131931 | 0.10 |
ENST00000547588.1
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr8_-_145047688 | 0.10 |
ENST00000356346.3
|
PLEC
|
plectin |
| chr11_+_64001962 | 0.10 |
ENST00000309422.2
|
VEGFB
|
vascular endothelial growth factor B |
| chr8_+_22844913 | 0.10 |
ENST00000519685.1
|
RHOBTB2
|
Rho-related BTB domain containing 2 |
| chr16_-_31085514 | 0.10 |
ENST00000300849.4
|
ZNF668
|
zinc finger protein 668 |
| chr14_+_24458021 | 0.10 |
ENST00000397071.1
ENST00000559411.1 ENST00000335125.6 |
DHRS4L2
|
dehydrogenase/reductase (SDR family) member 4 like 2 |
| chr3_-_48470838 | 0.10 |
ENST00000358459.4
ENST00000358536.4 |
PLXNB1
|
plexin B1 |
| chr5_-_176730676 | 0.09 |
ENST00000393611.2
ENST00000303251.6 ENST00000303270.6 |
RAB24
|
RAB24, member RAS oncogene family |
| chr8_+_22462532 | 0.09 |
ENST00000389279.3
|
CCAR2
|
cell cycle and apoptosis regulator 2 |
| chr20_+_2673383 | 0.09 |
ENST00000380648.4
ENST00000342725.5 |
EBF4
|
early B-cell factor 4 |
| chr3_-_49170522 | 0.09 |
ENST00000418109.1
|
LAMB2
|
laminin, beta 2 (laminin S) |
| chr16_-_1401799 | 0.09 |
ENST00000007390.2
|
TSR3
|
TSR3, 20S rRNA accumulation, homolog (S. cerevisiae) |
| chr20_-_44516256 | 0.09 |
ENST00000372519.3
|
SPATA25
|
spermatogenesis associated 25 |
| chr16_+_57126482 | 0.09 |
ENST00000537605.1
ENST00000535318.2 |
CPNE2
|
copine II |
| chr16_-_28503080 | 0.09 |
ENST00000565316.1
ENST00000565778.1 ENST00000357857.9 ENST00000568558.1 ENST00000357806.7 |
CLN3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr17_-_43502987 | 0.09 |
ENST00000376922.2
|
ARHGAP27
|
Rho GTPase activating protein 27 |
| chr11_-_417388 | 0.09 |
ENST00000332725.3
|
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr17_+_46985731 | 0.09 |
ENST00000360943.5
|
UBE2Z
|
ubiquitin-conjugating enzyme E2Z |
| chr16_-_71323617 | 0.09 |
ENST00000563876.1
|
CMTR2
|
cap methyltransferase 2 |
| chr22_+_42229100 | 0.09 |
ENST00000361204.4
|
SREBF2
|
sterol regulatory element binding transcription factor 2 |
| chr11_-_6502534 | 0.09 |
ENST00000254584.2
ENST00000525235.1 ENST00000445086.2 |
ARFIP2
|
ADP-ribosylation factor interacting protein 2 |
| chr14_+_92980111 | 0.09 |
ENST00000216487.7
ENST00000557762.1 |
RIN3
|
Ras and Rab interactor 3 |
| chr1_+_154377669 | 0.09 |
ENST00000368485.3
ENST00000344086.4 |
IL6R
|
interleukin 6 receptor |
| chr15_+_29211570 | 0.08 |
ENST00000558804.1
|
APBA2
|
amyloid beta (A4) precursor protein-binding, family A, member 2 |
| chr22_+_31742875 | 0.08 |
ENST00000504184.2
|
AC005003.1
|
CDNA FLJ20464 fis, clone KAT06158; HCG1777549; Uncharacterized protein |
| chr6_+_143381594 | 0.08 |
ENST00000367601.4
|
AIG1
|
androgen-induced 1 |
| chr4_+_8271471 | 0.08 |
ENST00000307358.2
ENST00000382512.3 |
HTRA3
|
HtrA serine peptidase 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TFAP2B
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0052151 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.1 | 0.8 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 0.3 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.1 | 0.7 | GO:0070235 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.2 | GO:0060490 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.1 | 0.2 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.1 | 0.2 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.0 | 0.3 | GO:0019087 | transformation of host cell by virus(GO:0019087) renal water absorption(GO:0070295) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.3 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.1 | GO:0061145 | bronchus cartilage development(GO:0060532) lung smooth muscle development(GO:0061145) |
| 0.0 | 0.2 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.3 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.2 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.1 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.1 | GO:0072312 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.0 | 0.2 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.3 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.0 | 0.1 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.1 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.2 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 0.1 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.0 | 0.1 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.0 | 0.1 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.0 | 0.2 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.1 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.0 | 0.1 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.1 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 0.1 | GO:0061760 | antifungal humoral response(GO:0019732) antifungal innate immune response(GO:0061760) |
| 0.0 | 0.1 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.1 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 | 0.1 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.0 | 0.1 | GO:2001178 | mediator complex assembly(GO:0036034) regulation of mediator complex assembly(GO:2001176) positive regulation of mediator complex assembly(GO:2001178) |
| 0.0 | 0.9 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.0 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.1 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.0 | 0.2 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.0 | 0.2 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.1 | GO:1990785 | response to water-immersion restraint stress(GO:1990785) |
| 0.0 | 0.1 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 0.1 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.2 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.0 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.0 | 0.1 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.1 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.0 | 0.3 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.1 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.3 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 0.7 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.4 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.6 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.1 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 | 0.2 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.2 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.2 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.1 | GO:1901162 | primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.1 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.2 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.0 | GO:2000742 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.0 | 0.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:1901382 | chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.0 | 0.1 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.0 | 0.1 | GO:1903906 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 | 0.1 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.0 | 0.3 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.0 | 0.0 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.2 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.1 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.1 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.0 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.1 | 0.2 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.1 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.0 | 0.2 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.1 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.0 | 0.4 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.1 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.8 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.2 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.4 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.3 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 0.4 | GO:0004324 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.1 | 0.3 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.1 | 0.3 | GO:0033906 | protein tyrosine kinase inhibitor activity(GO:0030292) hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.3 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.4 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.0 | 0.1 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.0 | 0.2 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.1 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.0 | 0.1 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.1 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.0 | 0.4 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.2 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.2 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.6 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:0010736 | serum response element binding(GO:0010736) |
| 0.0 | 0.1 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.1 | GO:0008478 | pyridoxal kinase activity(GO:0008478) |
| 0.0 | 0.1 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.5 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.1 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.0 | 0.1 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.0 | 0.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.0 | 0.1 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.0 | 0.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.2 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0016213 | linoleoyl-CoA desaturase activity(GO:0016213) |
| 0.0 | 0.4 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.3 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.0 | GO:0030617 | transforming growth factor beta receptor, inhibitory cytoplasmic mediator activity(GO:0030617) |
| 0.0 | 0.3 | GO:0033613 | activating transcription factor binding(GO:0033613) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.1 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.4 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |