Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
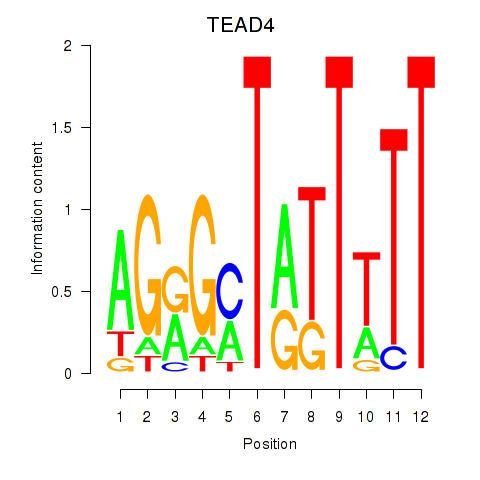
Results for TEAD4
Z-value: 0.44
Transcription factors associated with TEAD4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TEAD4
|
ENSG00000197905.4 | TEA domain transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TEAD4 | hg19_v2_chr12_+_3068544_3068597 | 0.95 | 4.6e-02 | Click! |
Activity profile of TEAD4 motif
Sorted Z-values of TEAD4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_37051798 | 0.26 |
ENST00000258829.5
|
NKX2-8
|
NK2 homeobox 8 |
| chr6_-_27835357 | 0.23 |
ENST00000331442.3
|
HIST1H1B
|
histone cluster 1, H1b |
| chr21_-_33975547 | 0.18 |
ENST00000431599.1
|
C21orf59
|
chromosome 21 open reading frame 59 |
| chr11_+_1295809 | 0.17 |
ENST00000598274.1
|
AC136297.1
|
Uncharacterized protein |
| chr7_+_13141010 | 0.17 |
ENST00000443947.1
|
AC011288.2
|
AC011288.2 |
| chr15_-_83224682 | 0.16 |
ENST00000562833.1
|
RP11-152F13.10
|
RP11-152F13.10 |
| chr22_-_46450024 | 0.15 |
ENST00000396008.2
ENST00000333761.1 |
C22orf26
|
chromosome 22 open reading frame 26 |
| chr16_-_69390209 | 0.14 |
ENST00000563634.1
|
RP11-343C2.9
|
Uncharacterized protein |
| chr12_-_121973974 | 0.14 |
ENST00000538379.1
ENST00000541318.1 ENST00000541511.1 |
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr22_-_42765174 | 0.13 |
ENST00000432473.1
ENST00000412060.1 ENST00000424852.1 |
Z83851.1
|
Z83851.1 |
| chr11_-_64570624 | 0.13 |
ENST00000439069.1
|
MAP4K2
|
mitogen-activated protein kinase kinase kinase kinase 2 |
| chr12_+_70574088 | 0.13 |
ENST00000552324.1
|
RP11-320P7.2
|
RP11-320P7.2 |
| chr19_+_9361606 | 0.13 |
ENST00000456448.1
|
OR7E24
|
olfactory receptor, family 7, subfamily E, member 24 |
| chr10_+_102758105 | 0.12 |
ENST00000429732.1
|
LZTS2
|
leucine zipper, putative tumor suppressor 2 |
| chr17_+_75396637 | 0.11 |
ENST00000590825.1
|
SEPT9
|
septin 9 |
| chr4_-_13546632 | 0.11 |
ENST00000382438.5
|
NKX3-2
|
NK3 homeobox 2 |
| chr2_+_196440692 | 0.11 |
ENST00000458054.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr9_+_131902283 | 0.11 |
ENST00000436883.1
ENST00000414510.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr20_-_3767324 | 0.11 |
ENST00000379751.4
|
CENPB
|
centromere protein B, 80kDa |
| chr19_+_9296279 | 0.11 |
ENST00000344248.2
|
OR7D2
|
olfactory receptor, family 7, subfamily D, member 2 |
| chr20_+_33814457 | 0.10 |
ENST00000246186.6
|
MMP24
|
matrix metallopeptidase 24 (membrane-inserted) |
| chr5_-_108063949 | 0.10 |
ENST00000606054.1
|
LINC01023
|
long intergenic non-protein coding RNA 1023 |
| chr4_+_96012585 | 0.10 |
ENST00000502683.1
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr7_-_37024665 | 0.10 |
ENST00000396040.2
|
ELMO1
|
engulfment and cell motility 1 |
| chr2_+_223916862 | 0.10 |
ENST00000604125.1
|
KCNE4
|
potassium voltage-gated channel, Isk-related family, member 4 |
| chr15_+_59908633 | 0.09 |
ENST00000559626.1
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr14_-_23451845 | 0.09 |
ENST00000262713.2
|
AJUBA
|
ajuba LIM protein |
| chr19_+_17403413 | 0.09 |
ENST00000595444.1
ENST00000600434.1 |
MRPL34
|
mitochondrial ribosomal protein L34 |
| chr16_-_73093597 | 0.09 |
ENST00000397992.5
|
ZFHX3
|
zinc finger homeobox 3 |
| chr3_+_149192475 | 0.09 |
ENST00000465758.1
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chrX_+_37639302 | 0.09 |
ENST00000545017.1
ENST00000536160.1 |
CYBB
|
cytochrome b-245, beta polypeptide |
| chr14_+_38033252 | 0.09 |
ENST00000554829.1
|
RP11-356O9.1
|
RP11-356O9.1 |
| chr12_+_28605426 | 0.09 |
ENST00000542801.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr12_+_117013656 | 0.09 |
ENST00000556529.1
|
MAP1LC3B2
|
microtubule-associated protein 1 light chain 3 beta 2 |
| chr16_+_4364762 | 0.09 |
ENST00000262366.3
|
GLIS2
|
GLIS family zinc finger 2 |
| chr16_-_2155399 | 0.08 |
ENST00000567946.1
|
PKD1
|
polycystic kidney disease 1 (autosomal dominant) |
| chr6_+_151358048 | 0.08 |
ENST00000450635.1
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr17_-_38545799 | 0.08 |
ENST00000577541.1
|
TOP2A
|
topoisomerase (DNA) II alpha 170kDa |
| chr2_+_38893208 | 0.08 |
ENST00000410063.1
|
GALM
|
galactose mutarotase (aldose 1-epimerase) |
| chr1_+_154229547 | 0.08 |
ENST00000428595.1
|
UBAP2L
|
ubiquitin associated protein 2-like |
| chr1_-_156252590 | 0.08 |
ENST00000361813.5
ENST00000368267.5 |
SMG5
|
SMG5 nonsense mediated mRNA decay factor |
| chr7_-_150777949 | 0.08 |
ENST00000482571.1
|
FASTK
|
Fas-activated serine/threonine kinase |
| chr17_+_17685422 | 0.07 |
ENST00000395774.1
|
RAI1
|
retinoic acid induced 1 |
| chrX_+_52240504 | 0.07 |
ENST00000399805.2
|
XAGE1B
|
X antigen family, member 1B |
| chr8_-_30002179 | 0.07 |
ENST00000320542.3
|
MBOAT4
|
membrane bound O-acyltransferase domain containing 4 |
| chr10_-_75410771 | 0.07 |
ENST00000372873.4
|
SYNPO2L
|
synaptopodin 2-like |
| chr12_-_111806892 | 0.07 |
ENST00000547710.1
ENST00000549321.1 ENST00000361483.3 ENST00000392658.5 |
FAM109A
|
family with sequence similarity 109, member A |
| chr3_-_12587055 | 0.07 |
ENST00000564146.3
|
C3orf83
|
chromosome 3 open reading frame 83 |
| chr12_+_56511943 | 0.07 |
ENST00000257940.2
ENST00000552345.1 ENST00000551880.1 ENST00000546903.1 ENST00000551790.1 |
ZC3H10
ESYT1
|
zinc finger CCCH-type containing 10 extended synaptotagmin-like protein 1 |
| chr2_-_231989808 | 0.07 |
ENST00000258400.3
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr9_-_33402506 | 0.07 |
ENST00000377425.4
ENST00000537089.1 ENST00000297988.1 ENST00000539936.1 ENST00000541274.1 |
AQP7
|
aquaporin 7 |
| chr3_+_111805262 | 0.07 |
ENST00000484828.1
|
C3orf52
|
chromosome 3 open reading frame 52 |
| chr1_+_171283331 | 0.07 |
ENST00000367749.3
|
FMO4
|
flavin containing monooxygenase 4 |
| chr6_-_13328564 | 0.06 |
ENST00000606530.1
ENST00000607658.1 ENST00000343141.4 ENST00000356436.4 ENST00000379300.3 ENST00000452989.1 ENST00000450347.1 ENST00000422136.1 ENST00000446018.1 ENST00000379291.1 ENST00000379307.2 ENST00000606370.1 ENST00000607230.1 |
TBC1D7
|
TBC1 domain family, member 7 |
| chr5_+_154092396 | 0.06 |
ENST00000336314.4
|
LARP1
|
La ribonucleoprotein domain family, member 1 |
| chr17_-_47865948 | 0.06 |
ENST00000513602.1
|
FAM117A
|
family with sequence similarity 117, member A |
| chr1_-_154928562 | 0.06 |
ENST00000368463.3
ENST00000539880.1 ENST00000542459.1 ENST00000368460.3 ENST00000368465.1 |
PBXIP1
|
pre-B-cell leukemia homeobox interacting protein 1 |
| chr9_+_131902346 | 0.06 |
ENST00000432124.1
ENST00000435305.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr18_+_3466248 | 0.06 |
ENST00000581029.1
ENST00000581442.1 ENST00000579007.1 |
RP11-838N2.4
|
RP11-838N2.4 |
| chr15_+_89164560 | 0.06 |
ENST00000379231.3
ENST00000559528.1 |
AEN
|
apoptosis enhancing nuclease |
| chr20_+_1246908 | 0.06 |
ENST00000381873.3
ENST00000381867.1 |
SNPH
|
syntaphilin |
| chr9_-_104249400 | 0.06 |
ENST00000374848.3
|
TMEM246
|
transmembrane protein 246 |
| chr5_+_174151536 | 0.05 |
ENST00000239243.6
ENST00000507785.1 |
MSX2
|
msh homeobox 2 |
| chr2_+_201987200 | 0.05 |
ENST00000425030.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr1_+_241695670 | 0.05 |
ENST00000366557.4
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr7_-_55606346 | 0.05 |
ENST00000545390.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr8_+_22225041 | 0.05 |
ENST00000289952.5
ENST00000524285.1 |
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr7_-_111428957 | 0.05 |
ENST00000417165.1
|
DOCK4
|
dedicator of cytokinesis 4 |
| chr4_+_144312659 | 0.05 |
ENST00000509992.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr1_-_158656488 | 0.05 |
ENST00000368147.4
|
SPTA1
|
spectrin, alpha, erythrocytic 1 (elliptocytosis 2) |
| chr17_+_4855053 | 0.05 |
ENST00000518175.1
|
ENO3
|
enolase 3 (beta, muscle) |
| chr19_+_35630628 | 0.05 |
ENST00000588715.1
ENST00000588607.1 |
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr6_-_25830785 | 0.05 |
ENST00000468082.1
|
SLC17A1
|
solute carrier family 17 (organic anion transporter), member 1 |
| chrX_-_52258669 | 0.05 |
ENST00000441417.1
|
XAGE1A
|
X antigen family, member 1A |
| chr10_+_5005445 | 0.05 |
ENST00000380872.4
|
AKR1C1
|
aldo-keto reductase family 1, member C1 |
| chr19_-_50083822 | 0.05 |
ENST00000596358.1
|
NOSIP
|
nitric oxide synthase interacting protein |
| chrX_+_37639264 | 0.05 |
ENST00000378588.4
|
CYBB
|
cytochrome b-245, beta polypeptide |
| chr12_+_10331605 | 0.05 |
ENST00000298530.3
|
TMEM52B
|
transmembrane protein 52B |
| chr1_+_46016703 | 0.05 |
ENST00000481885.1
ENST00000351829.4 ENST00000471651.1 |
AKR1A1
|
aldo-keto reductase family 1, member A1 (aldehyde reductase) |
| chr19_-_41256207 | 0.05 |
ENST00000598485.2
ENST00000470681.1 ENST00000339153.3 ENST00000598729.1 |
C19orf54
|
chromosome 19 open reading frame 54 |
| chr18_-_56985776 | 0.05 |
ENST00000587244.1
|
CPLX4
|
complexin 4 |
| chr20_+_62716348 | 0.05 |
ENST00000349451.3
|
OPRL1
|
opiate receptor-like 1 |
| chr11_-_6440624 | 0.05 |
ENST00000311051.3
|
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr20_+_61904556 | 0.05 |
ENST00000522403.2
ENST00000550188.1 |
ARFGAP1
|
ADP-ribosylation factor GTPase activating protein 1 |
| chr3_-_137834436 | 0.05 |
ENST00000327532.2
ENST00000467030.1 |
DZIP1L
|
DAZ interacting zinc finger protein 1-like |
| chr9_-_34397800 | 0.04 |
ENST00000297623.2
|
C9orf24
|
chromosome 9 open reading frame 24 |
| chr1_+_156252708 | 0.04 |
ENST00000295694.5
ENST00000357501.2 |
TMEM79
|
transmembrane protein 79 |
| chr11_-_59633951 | 0.04 |
ENST00000257264.3
|
TCN1
|
transcobalamin I (vitamin B12 binding protein, R binder family) |
| chr1_+_1215816 | 0.04 |
ENST00000379116.5
|
SCNN1D
|
sodium channel, non-voltage-gated 1, delta subunit |
| chr2_+_38893047 | 0.04 |
ENST00000272252.5
|
GALM
|
galactose mutarotase (aldose 1-epimerase) |
| chr8_+_107282368 | 0.04 |
ENST00000521369.2
|
RP11-395G23.3
|
RP11-395G23.3 |
| chr19_+_54369608 | 0.04 |
ENST00000336967.3
|
MYADM
|
myeloid-associated differentiation marker |
| chr16_+_29991673 | 0.04 |
ENST00000416441.2
|
TAOK2
|
TAO kinase 2 |
| chr14_-_77542485 | 0.04 |
ENST00000556781.1
ENST00000557526.1 ENST00000555512.1 |
RP11-7F17.3
|
RP11-7F17.3 |
| chr1_+_241695424 | 0.04 |
ENST00000366558.3
ENST00000366559.4 |
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr11_+_86502085 | 0.04 |
ENST00000527521.1
|
PRSS23
|
protease, serine, 23 |
| chr4_-_47983519 | 0.04 |
ENST00000358519.4
ENST00000544810.1 ENST00000402813.3 |
CNGA1
|
cyclic nucleotide gated channel alpha 1 |
| chr7_+_150811705 | 0.04 |
ENST00000335367.3
|
AGAP3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr21_+_17909594 | 0.04 |
ENST00000441820.1
ENST00000602280.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr21_-_27423339 | 0.04 |
ENST00000415997.1
|
APP
|
amyloid beta (A4) precursor protein |
| chr6_-_88411911 | 0.04 |
ENST00000257787.5
|
AKIRIN2
|
akirin 2 |
| chr10_-_735553 | 0.04 |
ENST00000280886.6
ENST00000423550.1 |
DIP2C
|
DIP2 disco-interacting protein 2 homolog C (Drosophila) |
| chr19_+_17862274 | 0.04 |
ENST00000596536.1
ENST00000593870.1 ENST00000598086.1 ENST00000598932.1 ENST00000595023.1 ENST00000594068.1 ENST00000596507.1 ENST00000595033.1 ENST00000597718.1 |
FCHO1
|
FCH domain only 1 |
| chrX_+_22056165 | 0.04 |
ENST00000535894.1
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked |
| chr4_+_186317133 | 0.04 |
ENST00000507753.1
|
ANKRD37
|
ankyrin repeat domain 37 |
| chr19_-_50083803 | 0.03 |
ENST00000391853.3
ENST00000339093.3 |
NOSIP
|
nitric oxide synthase interacting protein |
| chr10_-_4285923 | 0.03 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr20_+_43160458 | 0.03 |
ENST00000372889.1
ENST00000372887.1 ENST00000372882.3 |
PKIG
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr11_-_71639480 | 0.03 |
ENST00000529513.1
|
RP11-849H4.2
|
Putative short transient receptor potential channel 2-like protein |
| chr3_-_142608001 | 0.03 |
ENST00000295992.3
|
PCOLCE2
|
procollagen C-endopeptidase enhancer 2 |
| chr22_+_38142235 | 0.03 |
ENST00000407319.2
ENST00000403663.2 ENST00000428075.1 |
TRIOBP
|
TRIO and F-actin binding protein |
| chr8_+_24151553 | 0.03 |
ENST00000265769.4
ENST00000540823.1 ENST00000397649.3 |
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr1_+_169079823 | 0.03 |
ENST00000367813.3
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr4_+_75174180 | 0.03 |
ENST00000413830.1
|
EPGN
|
epithelial mitogen |
| chr1_+_114473350 | 0.03 |
ENST00000503968.1
|
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr4_-_110723194 | 0.03 |
ENST00000394635.3
|
CFI
|
complement factor I |
| chr3_-_69249863 | 0.03 |
ENST00000478263.1
ENST00000462512.1 |
FRMD4B
|
FERM domain containing 4B |
| chr1_-_207119738 | 0.03 |
ENST00000356495.4
|
PIGR
|
polymeric immunoglobulin receptor |
| chr3_+_186692745 | 0.03 |
ENST00000438590.1
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr1_+_16010779 | 0.03 |
ENST00000375799.3
ENST00000375793.2 |
PLEKHM2
|
pleckstrin homology domain containing, family M (with RUN domain) member 2 |
| chr4_-_159080806 | 0.03 |
ENST00000590648.1
|
FAM198B
|
family with sequence similarity 198, member B |
| chr7_-_43946175 | 0.03 |
ENST00000603700.1
ENST00000402306.3 ENST00000443736.1 ENST00000446958.1 |
URGCP-MRPS24
URGCP
|
URGCP-MRPS24 readthrough upregulator of cell proliferation |
| chr2_-_133429091 | 0.03 |
ENST00000345008.6
|
LYPD1
|
LY6/PLAUR domain containing 1 |
| chr15_+_80733570 | 0.03 |
ENST00000533983.1
ENST00000527771.1 ENST00000525103.1 |
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr12_-_125401885 | 0.03 |
ENST00000542416.1
|
UBC
|
ubiquitin C |
| chr7_+_23719749 | 0.03 |
ENST00000409192.3
ENST00000344962.4 ENST00000409653.1 ENST00000409994.3 |
FAM221A
|
family with sequence similarity 221, member A |
| chr1_+_244214577 | 0.03 |
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18 |
| chr7_-_41742697 | 0.03 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chr19_+_37837218 | 0.03 |
ENST00000591134.1
|
HKR1
|
HKR1, GLI-Kruppel zinc finger family member |
| chr20_+_3767547 | 0.03 |
ENST00000344256.6
ENST00000379598.5 |
CDC25B
|
cell division cycle 25B |
| chr6_-_152489484 | 0.03 |
ENST00000354674.4
ENST00000539504.1 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr8_+_55370487 | 0.03 |
ENST00000297316.4
|
SOX17
|
SRY (sex determining region Y)-box 17 |
| chr10_+_24528108 | 0.03 |
ENST00000438429.1
|
KIAA1217
|
KIAA1217 |
| chr5_-_41794313 | 0.03 |
ENST00000512084.1
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chr7_+_18535346 | 0.02 |
ENST00000405010.3
ENST00000406451.4 ENST00000428307.2 |
HDAC9
|
histone deacetylase 9 |
| chr7_+_13141097 | 0.02 |
ENST00000411542.1
|
AC011288.2
|
AC011288.2 |
| chr12_+_32260085 | 0.02 |
ENST00000548411.1
ENST00000281474.5 ENST00000551086.1 |
BICD1
|
bicaudal D homolog 1 (Drosophila) |
| chr17_+_57274914 | 0.02 |
ENST00000582004.1
ENST00000577660.1 |
PRR11
CTD-2510F5.6
|
proline rich 11 Uncharacterized protein |
| chr17_+_47865917 | 0.02 |
ENST00000259021.4
ENST00000454930.2 ENST00000509773.1 ENST00000510819.1 ENST00000424009.2 |
KAT7
|
K(lysine) acetyltransferase 7 |
| chr17_-_66951474 | 0.02 |
ENST00000269080.2
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr6_+_131958436 | 0.02 |
ENST00000357639.3
ENST00000543135.1 ENST00000427148.2 ENST00000358229.5 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chrX_-_55024967 | 0.02 |
ENST00000545676.1
|
PFKFB1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr3_-_196987309 | 0.02 |
ENST00000453607.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr17_-_66951382 | 0.02 |
ENST00000586539.1
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr15_-_72563585 | 0.02 |
ENST00000287196.9
ENST00000260376.7 |
PARP6
|
poly (ADP-ribose) polymerase family, member 6 |
| chr20_+_60174827 | 0.02 |
ENST00000543233.1
|
CDH4
|
cadherin 4, type 1, R-cadherin (retinal) |
| chr4_-_155511887 | 0.02 |
ENST00000302053.3
ENST00000403106.3 |
FGA
|
fibrinogen alpha chain |
| chr22_-_50523760 | 0.02 |
ENST00000395876.2
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr2_-_211341411 | 0.02 |
ENST00000233714.4
ENST00000443314.1 ENST00000441020.3 ENST00000450366.2 ENST00000431941.2 |
LANCL1
|
LanC lantibiotic synthetase component C-like 1 (bacterial) |
| chr21_+_47878757 | 0.02 |
ENST00000400274.1
ENST00000427143.2 ENST00000318711.7 ENST00000457905.3 ENST00000466639.1 ENST00000435722.3 ENST00000417564.2 |
DIP2A
|
DIP2 disco-interacting protein 2 homolog A (Drosophila) |
| chr15_+_81475047 | 0.02 |
ENST00000559388.1
|
IL16
|
interleukin 16 |
| chr4_+_96012614 | 0.02 |
ENST00000264568.4
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chrX_-_77150985 | 0.02 |
ENST00000358075.6
|
MAGT1
|
magnesium transporter 1 |
| chr1_+_154244987 | 0.02 |
ENST00000328703.7
ENST00000457918.2 ENST00000483970.2 ENST00000435087.1 ENST00000532105.1 |
HAX1
|
HCLS1 associated protein X-1 |
| chr11_+_12766583 | 0.02 |
ENST00000361985.2
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr4_-_48136217 | 0.02 |
ENST00000264316.4
|
TXK
|
TXK tyrosine kinase |
| chr11_-_71639613 | 0.02 |
ENST00000528184.1
ENST00000528511.2 |
RP11-849H4.2
|
Putative short transient receptor potential channel 2-like protein |
| chr5_-_32313019 | 0.02 |
ENST00000280285.5
ENST00000264934.5 |
MTMR12
|
myotubularin related protein 12 |
| chr10_+_63808970 | 0.02 |
ENST00000309334.5
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like) |
| chr12_-_111358372 | 0.02 |
ENST00000548438.1
ENST00000228841.8 |
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow |
| chr2_+_157330081 | 0.02 |
ENST00000409674.1
|
GPD2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr19_-_1650666 | 0.02 |
ENST00000588136.1
|
TCF3
|
transcription factor 3 |
| chr5_+_140261703 | 0.02 |
ENST00000409494.1
ENST00000289272.2 |
PCDHA13
|
protocadherin alpha 13 |
| chr6_+_125540951 | 0.02 |
ENST00000524679.1
|
TPD52L1
|
tumor protein D52-like 1 |
| chr11_-_71639446 | 0.02 |
ENST00000534704.1
|
RP11-849H4.2
|
Putative short transient receptor potential channel 2-like protein |
| chr9_+_90112117 | 0.02 |
ENST00000358077.5
|
DAPK1
|
death-associated protein kinase 1 |
| chr3_+_123813509 | 0.02 |
ENST00000460856.1
ENST00000240874.3 |
KALRN
|
kalirin, RhoGEF kinase |
| chr7_+_7196565 | 0.01 |
ENST00000429911.1
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr19_+_41698927 | 0.01 |
ENST00000310054.4
|
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1 |
| chr11_+_125496619 | 0.01 |
ENST00000532669.1
ENST00000278916.3 |
CHEK1
|
checkpoint kinase 1 |
| chr4_-_186732048 | 0.01 |
ENST00000448662.2
ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr2_-_37544209 | 0.01 |
ENST00000234179.2
|
PRKD3
|
protein kinase D3 |
| chr9_+_2029019 | 0.01 |
ENST00000382194.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr2_-_196933536 | 0.01 |
ENST00000312428.6
ENST00000410072.1 |
DNAH7
|
dynein, axonemal, heavy chain 7 |
| chr18_+_52258390 | 0.01 |
ENST00000321600.1
|
DYNAP
|
dynactin associated protein |
| chr21_-_44035168 | 0.01 |
ENST00000419628.1
|
AP001626.1
|
AP001626.1 |
| chr2_-_161056762 | 0.01 |
ENST00000428609.2
ENST00000409967.2 |
ITGB6
|
integrin, beta 6 |
| chr6_+_31638156 | 0.01 |
ENST00000409525.1
|
LY6G5B
|
lymphocyte antigen 6 complex, locus G5B |
| chr2_-_136875712 | 0.01 |
ENST00000241393.3
|
CXCR4
|
chemokine (C-X-C motif) receptor 4 |
| chr17_+_44588877 | 0.01 |
ENST00000576629.1
|
LRRC37A2
|
leucine rich repeat containing 37, member A2 |
| chr14_-_89883412 | 0.01 |
ENST00000557258.1
|
FOXN3
|
forkhead box N3 |
| chr2_+_145425573 | 0.01 |
ENST00000600064.1
ENST00000597670.1 ENST00000414256.1 ENST00000599187.1 ENST00000451774.1 ENST00000599072.1 ENST00000596589.1 ENST00000597893.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr22_-_50523688 | 0.01 |
ENST00000450140.2
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr14_+_68286478 | 0.01 |
ENST00000487861.1
|
RAD51B
|
RAD51 paralog B |
| chr4_-_110723134 | 0.01 |
ENST00000510800.1
ENST00000512148.1 |
CFI
|
complement factor I |
| chr7_+_18548924 | 0.01 |
ENST00000524023.1
|
HDAC9
|
histone deacetylase 9 |
| chrX_-_33146477 | 0.01 |
ENST00000378677.2
|
DMD
|
dystrophin |
| chr21_-_44299626 | 0.01 |
ENST00000330317.2
ENST00000398208.2 |
WDR4
|
WD repeat domain 4 |
| chr10_-_62149433 | 0.01 |
ENST00000280772.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr3_+_111805182 | 0.01 |
ENST00000430855.1
ENST00000431717.2 ENST00000264848.5 |
C3orf52
|
chromosome 3 open reading frame 52 |
| chr4_-_143226979 | 0.01 |
ENST00000514525.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr8_+_42128861 | 0.01 |
ENST00000518983.1
|
IKBKB
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta |
| chr2_+_152214098 | 0.01 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr1_+_206623784 | 0.01 |
ENST00000426388.1
|
SRGAP2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr18_-_25616519 | 0.01 |
ENST00000399380.3
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr19_+_19144384 | 0.01 |
ENST00000392335.2
ENST00000535612.1 ENST00000537263.1 ENST00000540707.1 ENST00000541725.1 ENST00000269932.6 ENST00000546344.1 ENST00000540792.1 ENST00000536098.1 ENST00000541898.1 ENST00000543877.1 |
ARMC6
|
armadillo repeat containing 6 |
| chr2_-_161056802 | 0.01 |
ENST00000283249.2
ENST00000409872.1 |
ITGB6
|
integrin, beta 6 |
| chr2_-_99279928 | 0.01 |
ENST00000414521.2
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr6_+_123317116 | 0.01 |
ENST00000275162.5
|
CLVS2
|
clavesin 2 |
| chr1_+_61547894 | 0.01 |
ENST00000403491.3
|
NFIA
|
nuclear factor I/A |
Network of associatons between targets according to the STRING database.
First level regulatory network of TEAD4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.1 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 0.1 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.1 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.1 | GO:1903613 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.0 | 0.1 | GO:0072287 | metanephric distal tubule morphogenesis(GO:0072287) |
| 0.0 | 0.2 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.0 | 0.1 | GO:0051795 | positive regulation of catagen(GO:0051795) activation of meiosis(GO:0090427) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.1 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.1 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.0 | 0.0 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0060994 | regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) |
| 0.0 | 0.1 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.0 | 0.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.1 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 0.1 | GO:0002133 | polycystin complex(GO:0002133) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.0 | 0.1 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.1 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.1 | GO:0047023 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) androsterone dehydrogenase activity(GO:0047023) indanol dehydrogenase activity(GO:0047718) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |