Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
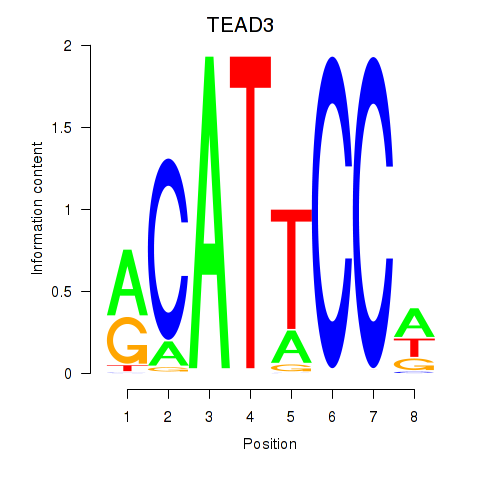
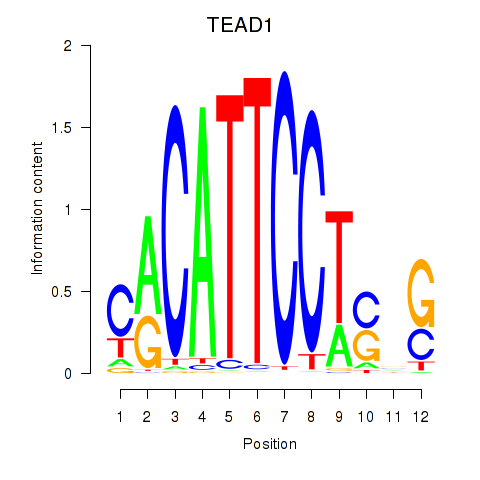
Results for TEAD3_TEAD1
Z-value: 0.56
Transcription factors associated with TEAD3_TEAD1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TEAD3
|
ENSG00000007866.14 | TEA domain transcription factor 3 |
|
TEAD1
|
ENSG00000187079.10 | TEA domain transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TEAD1 | hg19_v2_chr11_+_12695944_12695989 | 0.95 | 5.2e-02 | Click! |
| TEAD3 | hg19_v2_chr6_-_35464817_35464894 | 0.40 | 6.0e-01 | Click! |
Activity profile of TEAD3_TEAD1 motif
Sorted Z-values of TEAD3_TEAD1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_153279697 | 0.40 |
ENST00000444254.1
|
IRAK1
|
interleukin-1 receptor-associated kinase 1 |
| chr14_+_105559784 | 0.39 |
ENST00000548104.1
|
RP11-44N21.1
|
RP11-44N21.1 |
| chr12_-_12419905 | 0.39 |
ENST00000535731.1
|
LRP6
|
low density lipoprotein receptor-related protein 6 |
| chr11_-_62313090 | 0.38 |
ENST00000528508.1
ENST00000533365.1 |
AHNAK
|
AHNAK nucleoprotein |
| chr8_-_101322132 | 0.36 |
ENST00000523481.1
|
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr1_-_201438282 | 0.36 |
ENST00000367311.3
ENST00000367309.1 |
PHLDA3
|
pleckstrin homology-like domain, family A, member 3 |
| chr8_+_94752349 | 0.35 |
ENST00000391680.1
|
RBM12B-AS1
|
RBM12B antisense RNA 1 |
| chr19_+_34972543 | 0.34 |
ENST00000590071.2
|
WTIP
|
Wilms tumor 1 interacting protein |
| chr12_-_122241812 | 0.32 |
ENST00000538335.1
|
AC084018.1
|
AC084018.1 |
| chr4_+_76649753 | 0.30 |
ENST00000603759.1
|
USO1
|
USO1 vesicle transport factor |
| chr6_+_30850697 | 0.29 |
ENST00000509639.1
ENST00000412274.2 ENST00000507901.1 ENST00000507046.1 ENST00000437124.2 ENST00000454612.2 ENST00000396342.2 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr8_-_94753202 | 0.29 |
ENST00000521947.1
|
RBM12B
|
RNA binding motif protein 12B |
| chr7_+_100464760 | 0.26 |
ENST00000200457.4
|
TRIP6
|
thyroid hormone receptor interactor 6 |
| chr3_-_134093738 | 0.24 |
ENST00000506107.1
|
AMOTL2
|
angiomotin like 2 |
| chr11_+_66824303 | 0.22 |
ENST00000533360.1
|
RHOD
|
ras homolog family member D |
| chr8_-_144815966 | 0.21 |
ENST00000388913.3
|
FAM83H
|
family with sequence similarity 83, member H |
| chr9_-_14322319 | 0.20 |
ENST00000606230.1
|
NFIB
|
nuclear factor I/B |
| chr8_-_101321584 | 0.20 |
ENST00000523167.1
|
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr11_+_66824276 | 0.19 |
ENST00000308831.2
|
RHOD
|
ras homolog family member D |
| chr4_-_40632140 | 0.19 |
ENST00000514782.1
|
RBM47
|
RNA binding motif protein 47 |
| chr16_+_29823427 | 0.18 |
ENST00000358758.7
ENST00000567659.1 ENST00000572820.1 |
PRRT2
|
proline-rich transmembrane protein 2 |
| chr16_+_14396121 | 0.17 |
ENST00000570945.1
|
RP11-65J21.3
|
RP11-65J21.3 |
| chr12_-_7281469 | 0.17 |
ENST00000542370.1
ENST00000266560.3 |
RBP5
|
retinol binding protein 5, cellular |
| chr2_-_153573965 | 0.17 |
ENST00000448428.1
|
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr11_+_66824346 | 0.17 |
ENST00000532559.1
|
RHOD
|
ras homolog family member D |
| chr14_-_23451845 | 0.17 |
ENST00000262713.2
|
AJUBA
|
ajuba LIM protein |
| chr19_-_46272462 | 0.17 |
ENST00000317578.6
|
SIX5
|
SIX homeobox 5 |
| chr17_-_58603482 | 0.16 |
ENST00000585368.1
|
APPBP2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chr8_-_94752946 | 0.16 |
ENST00000519109.1
|
RBM12B
|
RNA binding motif protein 12B |
| chr16_-_88851618 | 0.16 |
ENST00000301015.9
|
PIEZO1
|
piezo-type mechanosensitive ion channel component 1 |
| chr6_-_42110342 | 0.16 |
ENST00000356542.5
|
C6orf132
|
chromosome 6 open reading frame 132 |
| chr12_-_53473136 | 0.15 |
ENST00000547837.1
ENST00000301463.4 |
SPRYD3
|
SPRY domain containing 3 |
| chr1_-_1293904 | 0.15 |
ENST00000309212.6
ENST00000342753.4 ENST00000445648.2 |
MXRA8
|
matrix-remodelling associated 8 |
| chr9_-_130700080 | 0.14 |
ENST00000373110.4
|
DPM2
|
dolichyl-phosphate mannosyltransferase polypeptide 2, regulatory subunit |
| chr14_-_23451467 | 0.14 |
ENST00000555074.1
ENST00000361265.4 |
RP11-298I3.5
AJUBA
|
RP11-298I3.5 ajuba LIM protein |
| chr2_-_161264385 | 0.14 |
ENST00000409972.1
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1 |
| chr7_-_2349579 | 0.14 |
ENST00000435060.1
|
SNX8
|
sorting nexin 8 |
| chr12_+_57916466 | 0.14 |
ENST00000355673.3
|
MBD6
|
methyl-CpG binding domain protein 6 |
| chr11_+_48002279 | 0.14 |
ENST00000534219.1
ENST00000527952.1 |
PTPRJ
|
protein tyrosine phosphatase, receptor type, J |
| chr8_-_94753184 | 0.13 |
ENST00000520560.1
|
RBM12B
|
RNA binding motif protein 12B |
| chr1_-_153935791 | 0.13 |
ENST00000429040.1
|
SLC39A1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr17_-_40575535 | 0.13 |
ENST00000357037.5
|
PTRF
|
polymerase I and transcript release factor |
| chr17_-_27949911 | 0.13 |
ENST00000492276.2
ENST00000345068.5 ENST00000584602.1 |
CORO6
|
coronin 6 |
| chr16_+_29823552 | 0.13 |
ENST00000300797.6
|
PRRT2
|
proline-rich transmembrane protein 2 |
| chr12_-_7245018 | 0.13 |
ENST00000543835.1
ENST00000535233.2 |
C1R
|
complement component 1, r subcomponent |
| chr6_-_34524093 | 0.12 |
ENST00000544425.1
|
SPDEF
|
SAM pointed domain containing ETS transcription factor |
| chr19_+_782755 | 0.12 |
ENST00000606242.1
ENST00000586061.1 |
AC006273.5
|
AC006273.5 |
| chr7_-_100881109 | 0.12 |
ENST00000308344.5
|
CLDN15
|
claudin 15 |
| chr8_+_21906433 | 0.12 |
ENST00000522148.1
|
DMTN
|
dematin actin binding protein |
| chr6_-_41909466 | 0.11 |
ENST00000414200.2
|
CCND3
|
cyclin D3 |
| chr19_-_14606900 | 0.11 |
ENST00000393029.3
ENST00000393028.1 ENST00000393033.4 ENST00000345425.2 ENST00000586027.1 ENST00000591349.1 ENST00000587210.1 |
GIPC1
|
GIPC PDZ domain containing family, member 1 |
| chr16_-_68002456 | 0.11 |
ENST00000576616.1
ENST00000572037.1 ENST00000338335.3 ENST00000422611.2 ENST00000316341.3 |
SLC12A4
|
solute carrier family 12 (potassium/chloride transporter), member 4 |
| chr14_-_77495007 | 0.11 |
ENST00000238647.3
|
IRF2BPL
|
interferon regulatory factor 2 binding protein-like |
| chr12_-_123565834 | 0.11 |
ENST00000546049.1
|
PITPNM2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chr1_-_25291475 | 0.11 |
ENST00000338888.3
ENST00000399916.1 |
RUNX3
|
runt-related transcription factor 3 |
| chr19_+_676385 | 0.11 |
ENST00000166139.4
|
FSTL3
|
follistatin-like 3 (secreted glycoprotein) |
| chr5_+_150406527 | 0.11 |
ENST00000520059.1
|
GPX3
|
glutathione peroxidase 3 (plasma) |
| chr22_+_37959647 | 0.11 |
ENST00000415670.1
|
CDC42EP1
|
CDC42 effector protein (Rho GTPase binding) 1 |
| chr14_+_76044934 | 0.11 |
ENST00000238667.4
|
FLVCR2
|
feline leukemia virus subgroup C cellular receptor family, member 2 |
| chr3_-_112360116 | 0.11 |
ENST00000206423.3
ENST00000439685.2 |
CCDC80
|
coiled-coil domain containing 80 |
| chr17_-_56492989 | 0.10 |
ENST00000583753.1
|
RNF43
|
ring finger protein 43 |
| chrX_+_70316005 | 0.10 |
ENST00000374259.3
|
FOXO4
|
forkhead box O4 |
| chr17_-_21156578 | 0.10 |
ENST00000399011.2
ENST00000468196.1 |
C17orf103
|
chromosome 17 open reading frame 103 |
| chr6_-_41909561 | 0.10 |
ENST00000372991.4
|
CCND3
|
cyclin D3 |
| chr14_+_65171099 | 0.10 |
ENST00000247226.7
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr6_+_30850862 | 0.10 |
ENST00000504651.1
ENST00000512694.1 ENST00000515233.1 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr12_-_49453557 | 0.10 |
ENST00000547610.1
|
KMT2D
|
lysine (K)-specific methyltransferase 2D |
| chr1_-_153935738 | 0.10 |
ENST00000417348.1
|
SLC39A1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr1_-_201096312 | 0.09 |
ENST00000449188.2
|
ASCL5
|
achaete-scute family bHLH transcription factor 5 |
| chr14_+_74035763 | 0.09 |
ENST00000238651.5
|
ACOT2
|
acyl-CoA thioesterase 2 |
| chr6_-_34524049 | 0.09 |
ENST00000374037.3
|
SPDEF
|
SAM pointed domain containing ETS transcription factor |
| chr16_-_122619 | 0.09 |
ENST00000262316.6
|
RHBDF1
|
rhomboid 5 homolog 1 (Drosophila) |
| chr7_-_100881041 | 0.09 |
ENST00000412417.1
ENST00000414035.1 |
CLDN15
|
claudin 15 |
| chr17_+_7338737 | 0.09 |
ENST00000323206.1
ENST00000396568.1 |
TMEM102
|
transmembrane protein 102 |
| chr11_+_65190245 | 0.09 |
ENST00000499732.1
ENST00000501122.2 ENST00000601801.1 |
NEAT1
|
nuclear paraspeckle assembly transcript 1 (non-protein coding) |
| chr19_+_15218180 | 0.09 |
ENST00000342784.2
ENST00000597977.1 ENST00000600440.1 |
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans) |
| chr2_-_190044480 | 0.09 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr17_-_73511584 | 0.09 |
ENST00000321617.3
|
CASKIN2
|
CASK interacting protein 2 |
| chr5_+_49962495 | 0.09 |
ENST00000515175.1
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr7_+_150725510 | 0.09 |
ENST00000461373.1
ENST00000358849.4 ENST00000297504.6 ENST00000542328.1 ENST00000498578.1 ENST00000356058.4 ENST00000477719.1 ENST00000477092.1 |
ABCB8
|
ATP-binding cassette, sub-family B (MDR/TAP), member 8 |
| chr1_+_154401791 | 0.08 |
ENST00000476006.1
|
IL6R
|
interleukin 6 receptor |
| chr4_+_129732419 | 0.08 |
ENST00000510308.1
|
PHF17
|
jade family PHD finger 1 |
| chr17_+_7255208 | 0.08 |
ENST00000333751.3
|
KCTD11
|
potassium channel tetramerization domain containing 11 |
| chr10_-_90712520 | 0.08 |
ENST00000224784.6
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr1_-_3447967 | 0.08 |
ENST00000294599.4
|
MEGF6
|
multiple EGF-like-domains 6 |
| chr4_-_40631859 | 0.08 |
ENST00000295971.7
ENST00000319592.4 |
RBM47
|
RNA binding motif protein 47 |
| chr7_-_135612198 | 0.08 |
ENST00000589735.1
|
LUZP6
|
leucine zipper protein 6 |
| chr11_-_8832521 | 0.08 |
ENST00000530438.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr12_-_498415 | 0.08 |
ENST00000535014.1
ENST00000543507.1 ENST00000544760.1 |
KDM5A
|
lysine (K)-specific demethylase 5A |
| chr11_-_85430356 | 0.08 |
ENST00000526999.1
|
SYTL2
|
synaptotagmin-like 2 |
| chr6_+_108881012 | 0.08 |
ENST00000343882.6
|
FOXO3
|
forkhead box O3 |
| chr17_-_7145106 | 0.08 |
ENST00000577035.1
|
GABARAP
|
GABA(A) receptor-associated protein |
| chrX_+_10031499 | 0.08 |
ENST00000454666.1
|
WWC3
|
WWC family member 3 |
| chr7_-_155604967 | 0.08 |
ENST00000297261.2
|
SHH
|
sonic hedgehog |
| chrX_+_99899180 | 0.08 |
ENST00000373004.3
|
SRPX2
|
sushi-repeat containing protein, X-linked 2 |
| chr1_+_200860122 | 0.08 |
ENST00000532631.1
ENST00000451872.2 |
C1orf106
|
chromosome 1 open reading frame 106 |
| chr16_-_31076273 | 0.07 |
ENST00000426488.2
|
ZNF668
|
zinc finger protein 668 |
| chr2_+_27505260 | 0.07 |
ENST00000380075.2
ENST00000296098.4 |
TRIM54
|
tripartite motif containing 54 |
| chr9_+_118916082 | 0.07 |
ENST00000328252.3
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1 |
| chr7_-_137686791 | 0.07 |
ENST00000452463.1
ENST00000330387.6 ENST00000456390.1 |
CREB3L2
|
cAMP responsive element binding protein 3-like 2 |
| chr14_-_21567009 | 0.07 |
ENST00000556174.1
ENST00000554478.1 ENST00000553980.1 ENST00000421093.2 |
ZNF219
|
zinc finger protein 219 |
| chr17_-_7297833 | 0.07 |
ENST00000571802.1
ENST00000576201.1 ENST00000573213.1 ENST00000324822.11 |
TMEM256-PLSCR3
|
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr3_-_127541194 | 0.07 |
ENST00000453507.2
|
MGLL
|
monoglyceride lipase |
| chr16_-_57514277 | 0.07 |
ENST00000562008.1
ENST00000567214.1 |
DOK4
|
docking protein 4 |
| chr17_-_73840774 | 0.07 |
ENST00000207549.4
|
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr1_+_12538594 | 0.07 |
ENST00000543710.1
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr5_+_176514413 | 0.07 |
ENST00000513166.1
|
FGFR4
|
fibroblast growth factor receptor 4 |
| chr12_+_14561422 | 0.07 |
ENST00000541056.1
|
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr19_-_55580838 | 0.07 |
ENST00000396247.3
ENST00000586945.1 ENST00000587026.1 |
RDH13
|
retinol dehydrogenase 13 (all-trans/9-cis) |
| chr15_+_42565393 | 0.07 |
ENST00000561871.1
|
GANC
|
glucosidase, alpha; neutral C |
| chr10_-_17659234 | 0.07 |
ENST00000466335.1
|
PTPLA
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A |
| chr3_-_9994021 | 0.07 |
ENST00000411976.2
ENST00000412055.1 |
PRRT3
|
proline-rich transmembrane protein 3 |
| chr3_-_134092561 | 0.07 |
ENST00000510560.1
ENST00000504234.1 ENST00000515172.1 |
AMOTL2
|
angiomotin like 2 |
| chr19_+_13906250 | 0.07 |
ENST00000254323.2
|
ZSWIM4
|
zinc finger, SWIM-type containing 4 |
| chr2_+_208104351 | 0.07 |
ENST00000440326.1
|
AC007879.7
|
AC007879.7 |
| chr3_+_49058444 | 0.07 |
ENST00000326925.6
ENST00000395458.2 |
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chrX_-_53310791 | 0.07 |
ENST00000375365.2
|
IQSEC2
|
IQ motif and Sec7 domain 2 |
| chr14_+_65170820 | 0.07 |
ENST00000555982.1
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr16_-_71264558 | 0.07 |
ENST00000448089.2
ENST00000393550.2 ENST00000448691.1 ENST00000393567.2 ENST00000321489.5 ENST00000539973.1 ENST00000288168.10 ENST00000545267.1 ENST00000541601.1 ENST00000538248.1 |
HYDIN
|
HYDIN, axonemal central pair apparatus protein |
| chr7_-_27219849 | 0.06 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr12_+_22778291 | 0.06 |
ENST00000545979.1
|
ETNK1
|
ethanolamine kinase 1 |
| chr14_+_35591020 | 0.06 |
ENST00000603611.1
|
KIAA0391
|
KIAA0391 |
| chr5_-_38845812 | 0.06 |
ENST00000513480.1
ENST00000512519.1 |
CTD-2127H9.1
|
CTD-2127H9.1 |
| chr12_-_7245152 | 0.06 |
ENST00000542220.2
|
C1R
|
complement component 1, r subcomponent |
| chr19_+_33182823 | 0.06 |
ENST00000397061.3
|
NUDT19
|
nudix (nucleoside diphosphate linked moiety X)-type motif 19 |
| chr12_+_57916584 | 0.06 |
ENST00000546632.1
ENST00000549623.1 ENST00000431731.2 |
MBD6
|
methyl-CpG binding domain protein 6 |
| chr11_-_1785139 | 0.06 |
ENST00000236671.2
|
CTSD
|
cathepsin D |
| chr12_-_109797249 | 0.06 |
ENST00000538041.1
|
RP11-256L11.1
|
RP11-256L11.1 |
| chr17_-_73505961 | 0.06 |
ENST00000433559.2
|
CASKIN2
|
CASK interacting protein 2 |
| chr12_-_498620 | 0.06 |
ENST00000399788.2
ENST00000382815.4 |
KDM5A
|
lysine (K)-specific demethylase 5A |
| chr1_+_78354243 | 0.06 |
ENST00000294624.8
|
NEXN
|
nexilin (F actin binding protein) |
| chr6_-_41909191 | 0.06 |
ENST00000512426.1
ENST00000372987.4 |
CCND3
|
cyclin D3 |
| chr5_+_140044261 | 0.06 |
ENST00000358337.5
|
WDR55
|
WD repeat domain 55 |
| chr1_+_18807424 | 0.06 |
ENST00000400664.1
|
KLHDC7A
|
kelch domain containing 7A |
| chr12_-_118490217 | 0.06 |
ENST00000542304.1
|
WSB2
|
WD repeat and SOCS box containing 2 |
| chr2_-_109605663 | 0.06 |
ENST00000409271.1
ENST00000258443.2 ENST00000376651.1 |
EDAR
|
ectodysplasin A receptor |
| chr1_-_161279749 | 0.06 |
ENST00000533357.1
ENST00000360451.6 ENST00000336559.4 |
MPZ
|
myelin protein zero |
| chr22_-_37584321 | 0.06 |
ENST00000397110.2
ENST00000337843.2 |
C1QTNF6
|
C1q and tumor necrosis factor related protein 6 |
| chr1_+_74701062 | 0.06 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chr4_+_110834033 | 0.06 |
ENST00000509793.1
ENST00000265171.5 |
EGF
|
epidermal growth factor |
| chr6_+_43139037 | 0.06 |
ENST00000265354.4
|
SRF
|
serum response factor (c-fos serum response element-binding transcription factor) |
| chr16_-_31076332 | 0.06 |
ENST00000539836.3
ENST00000535577.1 ENST00000442862.2 |
ZNF668
|
zinc finger protein 668 |
| chr14_-_59950724 | 0.06 |
ENST00000481608.1
|
L3HYPDH
|
L-3-hydroxyproline dehydratase (trans-) |
| chr2_-_107503558 | 0.06 |
ENST00000361686.4
ENST00000409087.3 |
ST6GAL2
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2 |
| chr19_+_44037546 | 0.06 |
ENST00000601282.1
|
ZNF575
|
zinc finger protein 575 |
| chr10_+_104404644 | 0.06 |
ENST00000462202.2
|
TRIM8
|
tripartite motif containing 8 |
| chr14_+_65171315 | 0.06 |
ENST00000394691.1
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr15_-_44069513 | 0.06 |
ENST00000433927.1
|
ELL3
|
elongation factor RNA polymerase II-like 3 |
| chr8_+_87111059 | 0.06 |
ENST00000285393.3
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr7_+_26438187 | 0.06 |
ENST00000439120.1
ENST00000430548.1 ENST00000421862.1 ENST00000449537.1 ENST00000420774.1 ENST00000418758.2 |
AC004540.5
|
AC004540.5 |
| chr11_+_58390132 | 0.06 |
ENST00000361987.4
|
CNTF
|
ciliary neurotrophic factor |
| chr16_-_30125177 | 0.06 |
ENST00000406256.3
|
GDPD3
|
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr7_+_150758304 | 0.06 |
ENST00000482950.1
ENST00000463414.1 ENST00000310317.5 |
SLC4A2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr17_-_73663245 | 0.06 |
ENST00000584999.1
ENST00000317905.5 ENST00000420326.2 ENST00000340830.5 |
RECQL5
|
RecQ protein-like 5 |
| chr12_+_113587558 | 0.06 |
ENST00000335621.6
|
CCDC42B
|
coiled-coil domain containing 42B |
| chr19_+_46010674 | 0.06 |
ENST00000245932.6
ENST00000592139.1 ENST00000590603.1 |
VASP
|
vasodilator-stimulated phosphoprotein |
| chr5_-_54988448 | 0.06 |
ENST00000503817.1
ENST00000512595.1 |
SLC38A9
|
solute carrier family 38, member 9 |
| chr19_-_11494975 | 0.06 |
ENST00000222139.6
ENST00000592375.2 |
EPOR
|
erythropoietin receptor |
| chr11_-_46940074 | 0.06 |
ENST00000378623.1
ENST00000534404.1 |
LRP4
|
low density lipoprotein receptor-related protein 4 |
| chr4_+_165675269 | 0.06 |
ENST00000507311.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr5_-_149682447 | 0.06 |
ENST00000328668.7
|
ARSI
|
arylsulfatase family, member I |
| chr19_-_47288162 | 0.06 |
ENST00000594991.1
|
SLC1A5
|
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr17_+_9728828 | 0.06 |
ENST00000262441.5
|
GLP2R
|
glucagon-like peptide 2 receptor |
| chrX_+_47420516 | 0.06 |
ENST00000377045.4
ENST00000290277.6 ENST00000377039.2 |
ARAF
|
v-raf murine sarcoma 3611 viral oncogene homolog |
| chr17_+_36873677 | 0.05 |
ENST00000471200.1
|
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6 |
| chr2_+_145780739 | 0.05 |
ENST00000597173.1
ENST00000602108.1 ENST00000420472.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr15_+_41245160 | 0.05 |
ENST00000444189.2
ENST00000446533.3 |
CHAC1
|
ChaC, cation transport regulator homolog 1 (E. coli) |
| chr2_+_45878407 | 0.05 |
ENST00000421201.1
|
PRKCE
|
protein kinase C, epsilon |
| chr17_-_8079632 | 0.05 |
ENST00000431792.2
|
TMEM107
|
transmembrane protein 107 |
| chr19_-_47287990 | 0.05 |
ENST00000593713.1
ENST00000598022.1 ENST00000434726.2 |
SLC1A5
|
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr22_+_42665742 | 0.05 |
ENST00000332965.3
ENST00000415205.1 ENST00000446578.1 |
Z83851.3
|
Z83851.3 |
| chr2_+_105050794 | 0.05 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr22_+_41697520 | 0.05 |
ENST00000352645.4
|
ZC3H7B
|
zinc finger CCCH-type containing 7B |
| chr3_-_12587055 | 0.05 |
ENST00000564146.3
|
C3orf83
|
chromosome 3 open reading frame 83 |
| chr16_-_57570450 | 0.05 |
ENST00000258214.2
|
CCDC102A
|
coiled-coil domain containing 102A |
| chr7_-_150946015 | 0.05 |
ENST00000262188.8
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr7_+_137761199 | 0.05 |
ENST00000411726.2
|
AKR1D1
|
aldo-keto reductase family 1, member D1 |
| chr9_-_35754253 | 0.05 |
ENST00000436428.2
|
MSMP
|
microseminoprotein, prostate associated |
| chrX_+_48455866 | 0.05 |
ENST00000376729.5
ENST00000218056.5 |
WDR13
|
WD repeat domain 13 |
| chr1_+_12524965 | 0.05 |
ENST00000471923.1
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr11_+_48002076 | 0.05 |
ENST00000418331.2
ENST00000440289.2 |
PTPRJ
|
protein tyrosine phosphatase, receptor type, J |
| chr17_-_27467418 | 0.05 |
ENST00000528564.1
|
MYO18A
|
myosin XVIIIA |
| chr15_-_72767490 | 0.05 |
ENST00000565181.1
|
RP11-1007O24.3
|
RP11-1007O24.3 |
| chr9_+_113431059 | 0.05 |
ENST00000416899.2
|
MUSK
|
muscle, skeletal, receptor tyrosine kinase |
| chr15_+_42566384 | 0.05 |
ENST00000440615.2
ENST00000318010.8 |
GANC
|
glucosidase, alpha; neutral C |
| chr16_+_29819446 | 0.05 |
ENST00000568282.1
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr19_+_42388437 | 0.05 |
ENST00000378152.4
ENST00000337665.4 |
ARHGEF1
|
Rho guanine nucleotide exchange factor (GEF) 1 |
| chr14_-_23446900 | 0.05 |
ENST00000556731.1
|
AJUBA
|
ajuba LIM protein |
| chr11_+_92085262 | 0.05 |
ENST00000298047.6
ENST00000409404.2 ENST00000541502.1 |
FAT3
|
FAT atypical cadherin 3 |
| chr1_+_44870866 | 0.05 |
ENST00000355387.2
ENST00000361799.2 |
RNF220
|
ring finger protein 220 |
| chr12_-_12419703 | 0.05 |
ENST00000543091.1
ENST00000261349.4 |
LRP6
|
low density lipoprotein receptor-related protein 6 |
| chr17_-_73840415 | 0.05 |
ENST00000592386.1
ENST00000412096.2 ENST00000586147.1 |
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr17_-_73663168 | 0.05 |
ENST00000578201.1
ENST00000423245.2 |
RECQL5
|
RecQ protein-like 5 |
| chr3_-_47950745 | 0.05 |
ENST00000429422.1
|
MAP4
|
microtubule-associated protein 4 |
| chr2_+_220325977 | 0.05 |
ENST00000396686.1
ENST00000396689.2 |
SPEG
|
SPEG complex locus |
| chr1_+_100435986 | 0.05 |
ENST00000532693.1
|
SLC35A3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 |
| chr2_+_145780767 | 0.05 |
ENST00000599358.1
ENST00000596278.1 ENST00000596747.1 ENST00000608652.1 ENST00000609705.1 ENST00000608432.1 ENST00000596970.1 ENST00000602041.1 ENST00000601578.1 ENST00000596034.1 ENST00000414195.2 ENST00000594837.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr2_-_75788038 | 0.05 |
ENST00000393913.3
ENST00000410113.1 |
EVA1A
|
eva-1 homolog A (C. elegans) |
| chr17_+_37856214 | 0.05 |
ENST00000445658.2
|
ERBB2
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 |
| chr2_+_168725458 | 0.05 |
ENST00000392690.3
|
B3GALT1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TEAD3_TEAD1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0035261 | external genitalia morphogenesis(GO:0035261) |
| 0.1 | 0.2 | GO:1902723 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.1 | 0.2 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 0.1 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 0.3 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.7 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.1 | GO:0034721 | histone H3-K4 demethylation, trimethyl-H3-K4-specific(GO:0034721) |
| 0.0 | 0.2 | GO:0060481 | lung goblet cell differentiation(GO:0060480) lobar bronchus epithelium development(GO:0060481) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.2 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.1 | GO:0010585 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.0 | 0.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.2 | GO:1990785 | response to water-immersion restraint stress(GO:1990785) |
| 0.0 | 0.1 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.0 | 0.1 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.1 | GO:1903412 | response to bile acid(GO:1903412) |
| 0.0 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.1 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.4 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.1 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.0 | 0.3 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.1 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.0 | 0.1 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.0 | 0.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.1 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.0 | 0.3 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.0 | 0.1 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.4 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.0 | 0.0 | GO:0042245 | RNA repair(GO:0042245) |
| 0.0 | 0.1 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.1 | GO:0072658 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.1 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.0 | 0.1 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.0 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.0 | 0.2 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.0 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.0 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.0 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.1 | GO:1900222 | negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.0 | 0.1 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.1 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 | 0.1 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.7 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 | 0.0 | GO:0002870 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.0 | 0.1 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.1 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.1 | GO:0072144 | glomerular mesangial cell development(GO:0072144) |
| 0.0 | 0.1 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.1 | 0.4 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1904928 | coreceptor activity involved in canonical Wnt signaling pathway(GO:1904928) |
| 0.1 | 0.4 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.2 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.1 | GO:0050333 | thiamin-triphosphatase activity(GO:0050333) |
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.1 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.0 | 0.1 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0047661 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.0 | 0.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.4 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.1 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.1 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.1 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.0 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.0 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.0 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.0 | 0.1 | GO:0010736 | serum response element binding(GO:0010736) |
| 0.0 | 0.0 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.0 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.0 | 0.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |