Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
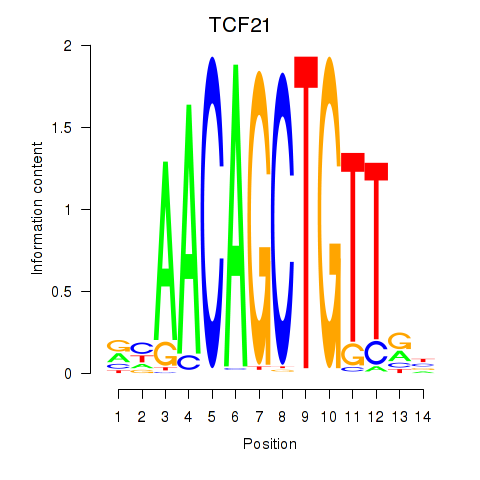
Results for TCF21
Z-value: 1.07
Transcription factors associated with TCF21
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TCF21
|
ENSG00000118526.6 | transcription factor 21 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TCF21 | hg19_v2_chr6_+_134210243_134210276 | 0.12 | 8.8e-01 | Click! |
Activity profile of TCF21 motif
Sorted Z-values of TCF21 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_68845417 | 0.92 |
ENST00000542875.1
|
RP11-81H14.2
|
RP11-81H14.2 |
| chr17_+_68165657 | 0.62 |
ENST00000243457.3
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr14_+_104710541 | 0.54 |
ENST00000419115.1
|
C14orf144
|
chromosome 14 open reading frame 144 |
| chr11_-_10920838 | 0.51 |
ENST00000503469.2
|
CTD-2003C8.2
|
CTD-2003C8.2 |
| chr11_+_117103441 | 0.46 |
ENST00000531287.1
ENST00000531452.1 |
RNF214
|
ring finger protein 214 |
| chr8_-_71519889 | 0.44 |
ENST00000521425.1
|
TRAM1
|
translocation associated membrane protein 1 |
| chr2_+_38177575 | 0.44 |
ENST00000407257.1
ENST00000417700.2 ENST00000234195.3 ENST00000442857.1 |
RMDN2
|
regulator of microtubule dynamics 2 |
| chr15_+_83776137 | 0.42 |
ENST00000322019.9
|
TM6SF1
|
transmembrane 6 superfamily member 1 |
| chr1_+_212738676 | 0.42 |
ENST00000366981.4
ENST00000366987.2 |
ATF3
|
activating transcription factor 3 |
| chr1_-_114429997 | 0.41 |
ENST00000471267.1
ENST00000393320.3 |
BCL2L15
|
BCL2-like 15 |
| chr6_-_31648150 | 0.39 |
ENST00000375858.3
ENST00000383237.4 |
LY6G5C
|
lymphocyte antigen 6 complex, locus G5C |
| chr2_-_122494487 | 0.38 |
ENST00000451734.1
ENST00000285814.4 |
MKI67IP
|
nucleolar protein interacting with the FHA domain of MKI67 |
| chr18_+_3247779 | 0.37 |
ENST00000578611.1
ENST00000583449.1 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr1_-_149889382 | 0.37 |
ENST00000369145.1
ENST00000369146.3 |
SV2A
|
synaptic vesicle glycoprotein 2A |
| chrX_+_15756382 | 0.37 |
ENST00000318636.3
|
CA5B
|
carbonic anhydrase VB, mitochondrial |
| chr3_-_156840776 | 0.36 |
ENST00000471357.1
|
LINC00880
|
long intergenic non-protein coding RNA 880 |
| chr2_+_33701684 | 0.36 |
ENST00000442390.1
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr8_-_99837856 | 0.35 |
ENST00000518165.1
ENST00000419617.2 |
STK3
|
serine/threonine kinase 3 |
| chr17_+_71228740 | 0.34 |
ENST00000268942.8
ENST00000359042.2 |
C17orf80
|
chromosome 17 open reading frame 80 |
| chr8_-_112248400 | 0.33 |
ENST00000519506.1
ENST00000522778.1 |
RP11-946L20.4
|
RP11-946L20.4 |
| chr15_+_75487984 | 0.32 |
ENST00000563905.1
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr5_+_49962772 | 0.32 |
ENST00000281631.5
ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr17_+_71229346 | 0.31 |
ENST00000535032.2
ENST00000582793.1 |
C17orf80
|
chromosome 17 open reading frame 80 |
| chr2_-_55276320 | 0.31 |
ENST00000357376.3
|
RTN4
|
reticulon 4 |
| chr12_+_16500599 | 0.30 |
ENST00000535309.1
ENST00000540056.1 ENST00000396209.1 ENST00000540126.1 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr17_+_71228793 | 0.30 |
ENST00000426147.2
|
C17orf80
|
chromosome 17 open reading frame 80 |
| chr7_+_99102573 | 0.29 |
ENST00000394170.2
|
ZKSCAN5
|
zinc finger with KRAB and SCAN domains 5 |
| chr14_-_53258180 | 0.28 |
ENST00000554230.1
|
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr6_-_74161977 | 0.28 |
ENST00000370318.1
ENST00000370315.3 |
MB21D1
|
Mab-21 domain containing 1 |
| chr12_-_11002063 | 0.28 |
ENST00000544994.1
ENST00000228811.4 ENST00000540107.1 |
PRR4
|
proline rich 4 (lacrimal) |
| chr1_-_150669604 | 0.28 |
ENST00000427665.1
ENST00000540514.1 |
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr8_+_33342268 | 0.27 |
ENST00000360128.6
|
MAK16
|
MAK16 homolog (S. cerevisiae) |
| chr7_+_66205712 | 0.27 |
ENST00000451741.2
ENST00000442563.1 ENST00000450873.2 ENST00000284957.5 |
KCTD7
RABGEF1
|
potassium channel tetramerization domain containing 7 RAB guanine nucleotide exchange factor (GEF) 1 |
| chr3_-_52869205 | 0.26 |
ENST00000446157.2
|
MUSTN1
|
musculoskeletal, embryonic nuclear protein 1 |
| chr1_+_78354175 | 0.26 |
ENST00000401035.3
ENST00000457030.1 ENST00000330010.8 |
NEXN
|
nexilin (F actin binding protein) |
| chr22_-_22863466 | 0.26 |
ENST00000406426.1
ENST00000360412.2 |
ZNF280B
|
zinc finger protein 280B |
| chr22_+_38071615 | 0.26 |
ENST00000215909.5
|
LGALS1
|
lectin, galactoside-binding, soluble, 1 |
| chr17_-_7493390 | 0.25 |
ENST00000538513.2
ENST00000570788.1 ENST00000250055.2 |
SOX15
|
SRY (sex determining region Y)-box 15 |
| chr2_+_113321939 | 0.25 |
ENST00000458012.2
|
POLR1B
|
polymerase (RNA) I polypeptide B, 128kDa |
| chr6_+_155470243 | 0.25 |
ENST00000456877.2
ENST00000528391.2 |
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr19_+_1452188 | 0.25 |
ENST00000587149.1
|
APC2
|
adenomatosis polyposis coli 2 |
| chr20_+_42839722 | 0.24 |
ENST00000442383.1
ENST00000435163.1 |
OSER1-AS1
|
OSER1 antisense RNA 1 (head to head) |
| chr17_+_16593539 | 0.24 |
ENST00000340621.5
ENST00000399273.1 ENST00000443444.2 ENST00000360524.8 ENST00000456009.1 |
CCDC144A
|
coiled-coil domain containing 144A |
| chr11_+_61722629 | 0.23 |
ENST00000526988.1
|
BEST1
|
bestrophin 1 |
| chrX_+_17755563 | 0.23 |
ENST00000380045.3
ENST00000380041.3 ENST00000380043.3 ENST00000398080.1 |
SCML1
|
sex comb on midleg-like 1 (Drosophila) |
| chr15_+_57998923 | 0.23 |
ENST00000380557.4
|
POLR2M
|
polymerase (RNA) II (DNA directed) polypeptide M |
| chr4_-_142253761 | 0.23 |
ENST00000511213.1
|
RP11-362F19.1
|
RP11-362F19.1 |
| chr17_-_62499334 | 0.22 |
ENST00000579996.1
|
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr19_-_33716750 | 0.22 |
ENST00000253188.4
|
SLC7A10
|
solute carrier family 7 (neutral amino acid transporter light chain, asc system), member 10 |
| chr4_+_106816592 | 0.22 |
ENST00000379987.2
ENST00000453617.2 ENST00000427316.2 ENST00000514622.1 ENST00000305572.8 |
NPNT
|
nephronectin |
| chr4_+_37892682 | 0.22 |
ENST00000508802.1
ENST00000261439.4 ENST00000402522.1 |
TBC1D1
|
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
| chr22_-_36924944 | 0.21 |
ENST00000405442.1
ENST00000402116.1 |
EIF3D
|
eukaryotic translation initiation factor 3, subunit D |
| chr4_-_186347099 | 0.21 |
ENST00000505357.1
ENST00000264689.6 |
UFSP2
|
UFM1-specific peptidase 2 |
| chr13_-_50510434 | 0.21 |
ENST00000361840.3
|
SPRYD7
|
SPRY domain containing 7 |
| chr7_+_99102267 | 0.21 |
ENST00000326775.5
ENST00000451158.1 |
ZKSCAN5
|
zinc finger with KRAB and SCAN domains 5 |
| chr11_+_71498552 | 0.21 |
ENST00000346333.6
ENST00000359244.4 ENST00000426628.2 |
FAM86C1
|
family with sequence similarity 86, member C1 |
| chr16_-_9030515 | 0.21 |
ENST00000535863.1
ENST00000381886.4 |
USP7
|
ubiquitin specific peptidase 7 (herpes virus-associated) |
| chr3_-_81792780 | 0.21 |
ENST00000489715.1
|
GBE1
|
glucan (1,4-alpha-), branching enzyme 1 |
| chr17_+_11924129 | 0.21 |
ENST00000353533.5
ENST00000415385.3 |
MAP2K4
|
mitogen-activated protein kinase kinase 4 |
| chr8_+_66955648 | 0.21 |
ENST00000522619.1
|
DNAJC5B
|
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
| chr12_+_122667658 | 0.20 |
ENST00000339777.4
ENST00000425921.1 |
LRRC43
|
leucine rich repeat containing 43 |
| chr2_+_101437487 | 0.20 |
ENST00000427413.1
ENST00000542504.1 |
NPAS2
|
neuronal PAS domain protein 2 |
| chr13_-_64650144 | 0.20 |
ENST00000456627.1
|
LINC00355
|
long intergenic non-protein coding RNA 355 |
| chr4_+_146403912 | 0.20 |
ENST00000507367.1
ENST00000394092.2 ENST00000515385.1 |
SMAD1
|
SMAD family member 1 |
| chr13_-_50510622 | 0.20 |
ENST00000378195.2
|
SPRYD7
|
SPRY domain containing 7 |
| chr4_-_4543700 | 0.20 |
ENST00000505286.1
ENST00000306200.2 |
STX18
|
syntaxin 18 |
| chr3_-_52868931 | 0.20 |
ENST00000486659.1
|
MUSTN1
|
musculoskeletal, embryonic nuclear protein 1 |
| chr20_-_52210368 | 0.19 |
ENST00000371471.2
|
ZNF217
|
zinc finger protein 217 |
| chr10_-_12084770 | 0.19 |
ENST00000357604.5
|
UPF2
|
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr4_+_164415594 | 0.19 |
ENST00000509657.1
ENST00000358572.5 |
TMA16
|
translation machinery associated 16 homolog (S. cerevisiae) |
| chr10_+_60094735 | 0.19 |
ENST00000373910.4
|
UBE2D1
|
ubiquitin-conjugating enzyme E2D 1 |
| chr12_+_9142131 | 0.18 |
ENST00000356986.3
ENST00000266551.4 |
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr17_+_29421987 | 0.18 |
ENST00000431387.4
|
NF1
|
neurofibromin 1 |
| chr22_-_23922410 | 0.18 |
ENST00000249053.3
|
IGLL1
|
immunoglobulin lambda-like polypeptide 1 |
| chr1_+_153600869 | 0.18 |
ENST00000292169.1
ENST00000368696.3 ENST00000436839.1 |
S100A1
|
S100 calcium binding protein A1 |
| chrX_-_107334790 | 0.18 |
ENST00000217958.3
|
PSMD10
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 |
| chr3_+_122296465 | 0.18 |
ENST00000483793.1
|
PARP15
|
poly (ADP-ribose) polymerase family, member 15 |
| chr4_+_186347388 | 0.17 |
ENST00000511138.1
ENST00000511581.1 |
C4orf47
|
chromosome 4 open reading frame 47 |
| chr14_+_58666824 | 0.17 |
ENST00000254286.4
|
ACTR10
|
actin-related protein 10 homolog (S. cerevisiae) |
| chr16_+_56970567 | 0.17 |
ENST00000563911.1
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr20_+_34802295 | 0.17 |
ENST00000432603.1
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr6_+_150070857 | 0.17 |
ENST00000544496.1
|
PCMT1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr20_-_3687775 | 0.17 |
ENST00000344754.4
ENST00000202578.4 |
SIGLEC1
|
sialic acid binding Ig-like lectin 1, sialoadhesin |
| chr3_-_105588231 | 0.17 |
ENST00000545639.1
ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr16_+_85936295 | 0.17 |
ENST00000563180.1
ENST00000564617.1 ENST00000564803.1 |
IRF8
|
interferon regulatory factor 8 |
| chr6_+_13182751 | 0.17 |
ENST00000415087.1
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr13_+_41635617 | 0.17 |
ENST00000542082.1
|
WBP4
|
WW domain binding protein 4 |
| chr21_+_37692481 | 0.16 |
ENST00000400485.1
|
MORC3
|
MORC family CW-type zinc finger 3 |
| chr2_+_177053307 | 0.16 |
ENST00000331462.4
|
HOXD1
|
homeobox D1 |
| chr19_+_11406821 | 0.16 |
ENST00000316737.1
ENST00000592955.1 ENST00000590327.1 |
TSPAN16
|
tetraspanin 16 |
| chr11_-_82997420 | 0.16 |
ENST00000455220.2
ENST00000529689.1 |
CCDC90B
|
coiled-coil domain containing 90B |
| chr2_-_234763105 | 0.16 |
ENST00000454020.1
|
HJURP
|
Holliday junction recognition protein |
| chr1_+_202976493 | 0.16 |
ENST00000367242.3
|
TMEM183A
|
transmembrane protein 183A |
| chr1_-_150669500 | 0.16 |
ENST00000271732.3
|
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr7_-_76255444 | 0.15 |
ENST00000454397.1
|
POMZP3
|
POM121 and ZP3 fusion |
| chr6_-_13711773 | 0.15 |
ENST00000011619.3
|
RANBP9
|
RAN binding protein 9 |
| chr1_+_78354330 | 0.15 |
ENST00000440324.1
|
NEXN
|
nexilin (F actin binding protein) |
| chrX_+_17755696 | 0.15 |
ENST00000419185.1
|
SCML1
|
sex comb on midleg-like 1 (Drosophila) |
| chr15_+_57998821 | 0.15 |
ENST00000299638.3
|
POLR2M
|
polymerase (RNA) II (DNA directed) polypeptide M |
| chr2_-_191115229 | 0.15 |
ENST00000409820.2
ENST00000410045.1 |
HIBCH
|
3-hydroxyisobutyryl-CoA hydrolase |
| chr8_-_141728760 | 0.15 |
ENST00000430260.2
|
PTK2
|
protein tyrosine kinase 2 |
| chr19_-_50528392 | 0.15 |
ENST00000600137.1
ENST00000597215.1 |
VRK3
|
vaccinia related kinase 3 |
| chr7_-_132766800 | 0.15 |
ENST00000542753.1
ENST00000448878.1 |
CHCHD3
|
coiled-coil-helix-coiled-coil-helix domain containing 3 |
| chr4_+_106816644 | 0.15 |
ENST00000506666.1
ENST00000503451.1 |
NPNT
|
nephronectin |
| chrX_-_108868390 | 0.15 |
ENST00000372101.2
|
KCNE1L
|
KCNE1-like |
| chr8_-_82633476 | 0.15 |
ENST00000518419.1
ENST00000517588.1 ENST00000521895.1 ENST00000520076.1 ENST00000519523.1 ENST00000522520.1 ENST00000521287.1 ENST00000523096.1 ENST00000220669.5 ENST00000517450.1 |
ZFAND1
|
zinc finger, AN1-type domain 1 |
| chr1_-_94374946 | 0.15 |
ENST00000370238.3
|
GCLM
|
glutamate-cysteine ligase, modifier subunit |
| chr11_+_47270436 | 0.15 |
ENST00000395397.3
ENST00000405576.1 |
NR1H3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr3_+_141106643 | 0.15 |
ENST00000514251.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr1_-_226076843 | 0.15 |
ENST00000272134.5
|
LEFTY1
|
left-right determination factor 1 |
| chr6_+_22221010 | 0.15 |
ENST00000567753.1
|
RP11-524C21.2
|
RP11-524C21.2 |
| chr2_-_17981462 | 0.15 |
ENST00000402989.1
ENST00000428868.1 |
SMC6
|
structural maintenance of chromosomes 6 |
| chr22_+_31277661 | 0.14 |
ENST00000454145.1
ENST00000453621.1 ENST00000431368.1 ENST00000535268.1 |
OSBP2
|
oxysterol binding protein 2 |
| chr10_+_52499682 | 0.14 |
ENST00000185907.9
ENST00000374006.1 |
ASAH2B
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2B |
| chr12_-_68845165 | 0.14 |
ENST00000360485.3
ENST00000441255.2 |
RP11-81H14.2
|
RP11-81H14.2 |
| chr14_-_45431091 | 0.14 |
ENST00000579157.1
ENST00000396128.4 ENST00000556500.1 |
KLHL28
|
kelch-like family member 28 |
| chr6_-_30080876 | 0.14 |
ENST00000376734.3
|
TRIM31
|
tripartite motif containing 31 |
| chr17_-_7081435 | 0.14 |
ENST00000380920.4
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr5_-_34043310 | 0.14 |
ENST00000231338.7
|
C1QTNF3
|
C1q and tumor necrosis factor related protein 3 |
| chr5_+_125967372 | 0.14 |
ENST00000357147.3
|
C5orf48
|
chromosome 5 open reading frame 48 |
| chr5_+_170288856 | 0.14 |
ENST00000523189.1
|
RANBP17
|
RAN binding protein 17 |
| chr18_-_55470320 | 0.14 |
ENST00000536015.1
|
ATP8B1
|
ATPase, aminophospholipid transporter, class I, type 8B, member 1 |
| chr18_+_158513 | 0.14 |
ENST00000400266.3
ENST00000580410.1 ENST00000383589.2 ENST00000261601.7 |
USP14
|
ubiquitin specific peptidase 14 (tRNA-guanine transglycosylase) |
| chr3_+_167453493 | 0.14 |
ENST00000295777.5
ENST00000472747.2 |
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr2_+_33701707 | 0.14 |
ENST00000425210.1
ENST00000444784.1 ENST00000423159.1 |
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr1_+_14075865 | 0.14 |
ENST00000413440.1
|
PRDM2
|
PR domain containing 2, with ZNF domain |
| chr10_-_28571015 | 0.14 |
ENST00000375719.3
ENST00000375732.1 |
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr14_+_31091511 | 0.14 |
ENST00000544052.2
ENST00000421551.3 ENST00000541123.1 ENST00000557076.1 ENST00000553693.1 ENST00000396629.2 |
SCFD1
|
sec1 family domain containing 1 |
| chrX_-_107334750 | 0.14 |
ENST00000340200.5
ENST00000372296.1 ENST00000372295.1 ENST00000361815.5 |
PSMD10
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 |
| chr19_-_43702231 | 0.14 |
ENST00000597374.1
ENST00000599371.1 |
PSG4
|
pregnancy specific beta-1-glycoprotein 4 |
| chr1_+_78354297 | 0.14 |
ENST00000334785.7
|
NEXN
|
nexilin (F actin binding protein) |
| chr18_+_56807096 | 0.14 |
ENST00000588875.1
|
SEC11C
|
SEC11 homolog C (S. cerevisiae) |
| chr11_+_7597639 | 0.13 |
ENST00000533792.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr6_+_143772060 | 0.13 |
ENST00000367591.4
|
PEX3
|
peroxisomal biogenesis factor 3 |
| chr3_+_145782358 | 0.13 |
ENST00000422482.1
|
AC107021.1
|
HCG1786590; PRO2533; Uncharacterized protein |
| chr12_-_52845910 | 0.13 |
ENST00000252252.3
|
KRT6B
|
keratin 6B |
| chr7_-_132766818 | 0.13 |
ENST00000262570.5
|
CHCHD3
|
coiled-coil-helix-coiled-coil-helix domain containing 3 |
| chr3_-_176915215 | 0.13 |
ENST00000457928.2
ENST00000422442.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr16_-_5147743 | 0.13 |
ENST00000587133.1
ENST00000458008.4 ENST00000427587.4 |
FAM86A
|
family with sequence similarity 86, member A |
| chr12_-_105352080 | 0.13 |
ENST00000433540.1
|
SLC41A2
|
solute carrier family 41 (magnesium transporter), member 2 |
| chr17_-_63052929 | 0.13 |
ENST00000439174.2
|
GNA13
|
guanine nucleotide binding protein (G protein), alpha 13 |
| chr22_+_32754139 | 0.13 |
ENST00000382088.3
|
RFPL3
|
ret finger protein-like 3 |
| chr1_+_25757376 | 0.13 |
ENST00000399766.3
ENST00000399763.3 ENST00000374343.4 |
TMEM57
|
transmembrane protein 57 |
| chr17_-_7193711 | 0.13 |
ENST00000571464.1
|
YBX2
|
Y box binding protein 2 |
| chr2_+_63816126 | 0.13 |
ENST00000454035.1
|
MDH1
|
malate dehydrogenase 1, NAD (soluble) |
| chr3_-_10332416 | 0.12 |
ENST00000450603.1
ENST00000449554.2 |
GHRL
|
ghrelin/obestatin prepropeptide |
| chr12_+_16500571 | 0.12 |
ENST00000543076.1
ENST00000396210.3 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr2_+_74361599 | 0.12 |
ENST00000401851.1
|
MGC10955
|
MGC10955 |
| chr22_+_44351419 | 0.12 |
ENST00000396202.3
|
SAMM50
|
SAMM50 sorting and assembly machinery component |
| chr3_-_105587879 | 0.12 |
ENST00000264122.4
ENST00000403724.1 ENST00000405772.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr15_+_63796779 | 0.12 |
ENST00000561442.1
ENST00000560070.1 ENST00000540797.1 ENST00000380324.3 ENST00000268049.7 ENST00000536001.1 ENST00000539772.1 |
USP3
|
ubiquitin specific peptidase 3 |
| chr11_-_18656028 | 0.12 |
ENST00000336349.5
|
SPTY2D1
|
SPT2, Suppressor of Ty, domain containing 1 (S. cerevisiae) |
| chr1_-_31230650 | 0.12 |
ENST00000294507.3
|
LAPTM5
|
lysosomal protein transmembrane 5 |
| chr3_+_179065474 | 0.12 |
ENST00000471841.1
ENST00000280653.7 |
MFN1
|
mitofusin 1 |
| chr2_-_75745823 | 0.12 |
ENST00000452003.1
|
EVA1A
|
eva-1 homolog A (C. elegans) |
| chr11_-_88070920 | 0.12 |
ENST00000524463.1
ENST00000227266.5 |
CTSC
|
cathepsin C |
| chr20_+_30327063 | 0.12 |
ENST00000300403.6
ENST00000340513.4 |
TPX2
|
TPX2, microtubule-associated |
| chr19_-_17185848 | 0.12 |
ENST00000593360.1
|
HAUS8
|
HAUS augmin-like complex, subunit 8 |
| chr12_-_10875831 | 0.12 |
ENST00000279550.7
ENST00000228251.4 |
YBX3
|
Y box binding protein 3 |
| chr3_+_169684553 | 0.12 |
ENST00000337002.4
ENST00000480708.1 |
SEC62
|
SEC62 homolog (S. cerevisiae) |
| chr11_-_62457371 | 0.12 |
ENST00000317449.4
|
LRRN4CL
|
LRRN4 C-terminal like |
| chr12_+_112563335 | 0.12 |
ENST00000549358.1
ENST00000257604.5 ENST00000548092.1 ENST00000552896.1 |
TRAFD1
|
TRAF-type zinc finger domain containing 1 |
| chr10_+_35415978 | 0.12 |
ENST00000429130.3
ENST00000469949.2 ENST00000460270.1 |
CREM
|
cAMP responsive element modulator |
| chr17_+_34848049 | 0.12 |
ENST00000588902.1
ENST00000591067.1 |
ZNHIT3
|
zinc finger, HIT-type containing 3 |
| chr16_+_16484691 | 0.12 |
ENST00000344087.4
|
NPIPA7
|
nuclear pore complex interacting protein family, member A7 |
| chr5_-_142784003 | 0.12 |
ENST00000416954.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr17_+_77681075 | 0.11 |
ENST00000397549.2
|
CTD-2116F7.1
|
CTD-2116F7.1 |
| chr16_-_11485922 | 0.11 |
ENST00000599216.1
|
CTD-3088G3.8
|
Protein LOC388210 |
| chrX_+_107334895 | 0.11 |
ENST00000372232.3
ENST00000345734.3 ENST00000372254.3 |
ATG4A
|
autophagy related 4A, cysteine peptidase |
| chr5_-_179050066 | 0.11 |
ENST00000329433.6
ENST00000510411.1 |
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr19_+_11651942 | 0.11 |
ENST00000587087.1
|
CNN1
|
calponin 1, basic, smooth muscle |
| chr11_-_64512803 | 0.11 |
ENST00000377489.1
ENST00000354024.3 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr6_-_41673552 | 0.11 |
ENST00000419574.1
ENST00000445214.1 |
TFEB
|
transcription factor EB |
| chr1_+_46859933 | 0.11 |
ENST00000243167.8
|
FAAH
|
fatty acid amide hydrolase |
| chr20_-_23030296 | 0.11 |
ENST00000377103.2
|
THBD
|
thrombomodulin |
| chrX_-_16887963 | 0.11 |
ENST00000380084.4
|
RBBP7
|
retinoblastoma binding protein 7 |
| chr17_-_39623681 | 0.11 |
ENST00000225899.3
|
KRT32
|
keratin 32 |
| chr18_+_21529811 | 0.11 |
ENST00000588004.1
|
LAMA3
|
laminin, alpha 3 |
| chr6_+_44184653 | 0.11 |
ENST00000573382.2
ENST00000576476.1 |
RP1-302G2.5
|
RP1-302G2.5 |
| chr8_+_19171128 | 0.11 |
ENST00000265807.3
|
SH2D4A
|
SH2 domain containing 4A |
| chr15_+_44084503 | 0.11 |
ENST00000409960.2
ENST00000409646.1 ENST00000594896.1 ENST00000339624.5 ENST00000409291.1 ENST00000402131.1 ENST00000403425.1 ENST00000430901.1 |
SERF2
|
small EDRK-rich factor 2 |
| chr11_+_6947720 | 0.11 |
ENST00000414517.2
|
ZNF215
|
zinc finger protein 215 |
| chr22_+_44351301 | 0.11 |
ENST00000350028.4
|
SAMM50
|
SAMM50 sorting and assembly machinery component |
| chr15_+_63340775 | 0.11 |
ENST00000559281.1
ENST00000317516.7 |
TPM1
|
tropomyosin 1 (alpha) |
| chr17_+_7462103 | 0.11 |
ENST00000396545.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr11_-_31531121 | 0.10 |
ENST00000532287.1
ENST00000526776.1 ENST00000534812.1 ENST00000529749.1 ENST00000278200.1 ENST00000530023.1 ENST00000533642.1 |
IMMP1L
|
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr16_+_18995249 | 0.10 |
ENST00000569532.1
ENST00000304381.5 |
TMC7
|
transmembrane channel-like 7 |
| chr2_+_231280954 | 0.10 |
ENST00000409824.1
ENST00000409341.1 ENST00000409112.1 ENST00000340126.4 ENST00000341950.4 |
SP100
|
SP100 nuclear antigen |
| chr17_+_29421900 | 0.10 |
ENST00000358273.4
ENST00000356175.3 |
NF1
|
neurofibromin 1 |
| chr21_-_10990830 | 0.10 |
ENST00000361285.4
ENST00000342420.5 ENST00000328758.5 |
TPTE
|
transmembrane phosphatase with tensin homology |
| chr6_+_31555045 | 0.10 |
ENST00000396101.3
ENST00000490742.1 |
LST1
|
leukocyte specific transcript 1 |
| chr7_-_6523755 | 0.10 |
ENST00000436575.1
ENST00000258739.4 |
DAGLB
KDELR2
|
diacylglycerol lipase, beta KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 |
| chr2_+_231280908 | 0.10 |
ENST00000427101.2
ENST00000432979.1 |
SP100
|
SP100 nuclear antigen |
| chr12_+_72233487 | 0.10 |
ENST00000482439.2
ENST00000550746.1 ENST00000491063.1 ENST00000319106.8 ENST00000485960.2 ENST00000393309.3 |
TBC1D15
|
TBC1 domain family, member 15 |
| chr15_+_45028520 | 0.10 |
ENST00000329464.4
|
TRIM69
|
tripartite motif containing 69 |
| chr6_+_150070831 | 0.10 |
ENST00000367380.5
|
PCMT1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr15_+_93447675 | 0.10 |
ENST00000536619.1
|
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr15_+_40987661 | 0.10 |
ENST00000526763.1
|
RAD51
|
RAD51 recombinase |
| chr17_+_7477040 | 0.10 |
ENST00000581384.1
ENST00000577929.1 |
EIF4A1
|
eukaryotic translation initiation factor 4A1 |
| chr7_+_87563557 | 0.10 |
ENST00000439864.1
ENST00000412441.1 ENST00000398201.4 ENST00000265727.7 ENST00000315984.7 ENST00000398209.3 |
ADAM22
|
ADAM metallopeptidase domain 22 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TCF21
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 0.4 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.1 | 0.4 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.1 | 0.4 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 0.1 | 0.4 | GO:0097195 | pilomotor reflex(GO:0097195) |
| 0.1 | 0.3 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.1 | 0.3 | GO:0036216 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.1 | 0.2 | GO:0048627 | myoblast development(GO:0048627) |
| 0.1 | 0.2 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.1 | 0.2 | GO:0031662 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) response to DDT(GO:0046680) histone H3-S10 phosphorylation involved in chromosome condensation(GO:2000775) |
| 0.1 | 0.4 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 0.3 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.1 | 0.2 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 | 0.4 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.1 | 0.2 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 | 0.3 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.1 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.0 | 0.0 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.0 | 0.2 | GO:0044752 | response to human chorionic gonadotropin(GO:0044752) |
| 0.0 | 0.2 | GO:2000672 | cellular response to sorbitol(GO:0072709) negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.1 | GO:1904344 | positive regulation of growth rate(GO:0040010) regulation of gastric mucosal blood circulation(GO:1904344) positive regulation of gastric mucosal blood circulation(GO:1904346) regulation of small intestine smooth muscle contraction(GO:1904347) positive regulation of small intestine smooth muscle contraction(GO:1904349) gastric mucosal blood circulation(GO:1990768) small intestine smooth muscle contraction(GO:1990770) |
| 0.0 | 0.3 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.1 | GO:0070781 | response to biotin(GO:0070781) |
| 0.0 | 0.2 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.0 | 0.3 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 | 0.4 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.1 | GO:1990426 | homologous recombination-dependent replication fork processing(GO:1990426) |
| 0.0 | 0.1 | GO:0071301 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.0 | 0.1 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.0 | 0.1 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.0 | 0.3 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.1 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 0.0 | 0.1 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.1 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) |
| 0.0 | 0.2 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.2 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.0 | 0.2 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.1 | GO:0072616 | interleukin-18 secretion(GO:0072616) |
| 0.0 | 0.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.2 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.1 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.0 | 0.1 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.0 | 0.1 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.1 | GO:1901656 | glycoside transport(GO:1901656) |
| 0.0 | 0.5 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.3 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.2 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.2 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.2 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.1 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.1 | GO:0051037 | regulation of transcription involved in meiotic cell cycle(GO:0051037) |
| 0.0 | 0.1 | GO:0043012 | regulation of fusion of sperm to egg plasma membrane(GO:0043012) |
| 0.0 | 0.1 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.0 | 0.4 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.2 | GO:0044146 | negative regulation of growth of symbiont in host(GO:0044130) negative regulation of growth of symbiont involved in interaction with host(GO:0044146) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.0 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.2 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.2 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.3 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.1 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.0 | GO:0032289 | central nervous system myelin formation(GO:0032289) detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:0060152 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 0.1 | GO:0071638 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 0.2 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.0 | 0.3 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.1 | GO:0001807 | regulation of type IV hypersensitivity(GO:0001807) negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.0 | 0.1 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.3 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.0 | 0.2 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.0 | 0.1 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.0 | 0.1 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.2 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.2 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.4 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.2 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.1 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.2 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.1 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 0.4 | GO:0034678 | smooth muscle contractile fiber(GO:0030485) integrin alpha8-beta1 complex(GO:0034678) |
| 0.1 | 0.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.1 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.2 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 0.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.2 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.3 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.1 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.4 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.0 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.3 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.3 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.4 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.0 | 0.1 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.0 | 0.2 | GO:0047860 | diiodophenylpyruvate reductase activity(GO:0047860) |
| 0.0 | 0.1 | GO:0031768 | growth hormone-releasing hormone activity(GO:0016608) ghrelin receptor binding(GO:0031768) |
| 0.0 | 0.6 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.3 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.1 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.0 | 0.1 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.0 | 0.1 | GO:0072591 | citrate-L-glutamate ligase activity(GO:0072591) |
| 0.0 | 0.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.1 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.1 | GO:0004040 | amidase activity(GO:0004040) |
| 0.0 | 0.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.1 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.1 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.3 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.1 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.6 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.4 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.1 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.0 | 0.0 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.0 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.2 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.5 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 0.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.4 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.0 | GO:0045155 | electron transporter, transferring electrons from CoQH2-cytochrome c reductase complex and cytochrome c oxidase complex activity(GO:0045155) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | PID RAS PATHWAY | Regulation of Ras family activation |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.6 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.2 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |