Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
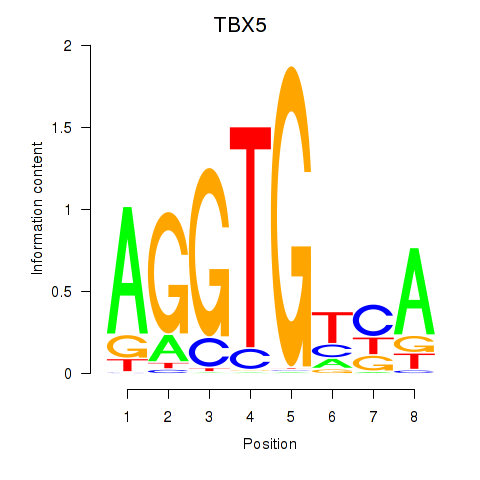
Results for TBX5
Z-value: 0.65
Transcription factors associated with TBX5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX5
|
ENSG00000089225.15 | T-box transcription factor 5 |
Activity profile of TBX5 motif
Sorted Z-values of TBX5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_64068116 | 0.33 |
ENST00000480679.1
|
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr15_+_90319557 | 0.32 |
ENST00000341735.3
|
MESP2
|
mesoderm posterior 2 homolog (mouse) |
| chr15_+_76030311 | 0.32 |
ENST00000543887.1
|
AC019294.1
|
AC019294.1 |
| chr5_-_100238918 | 0.25 |
ENST00000451528.2
|
ST8SIA4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 |
| chr3_-_9811674 | 0.23 |
ENST00000411972.1
|
CAMK1
|
calcium/calmodulin-dependent protein kinase I |
| chr4_-_102268708 | 0.22 |
ENST00000525819.1
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr14_-_37051798 | 0.21 |
ENST00000258829.5
|
NKX2-8
|
NK2 homeobox 8 |
| chr1_-_12679171 | 0.20 |
ENST00000606790.1
|
RP11-474O21.5
|
RP11-474O21.5 |
| chr21_+_44866471 | 0.19 |
ENST00000448049.1
|
LINC00319
|
long intergenic non-protein coding RNA 319 |
| chr8_-_99955042 | 0.19 |
ENST00000519420.1
|
STK3
|
serine/threonine kinase 3 |
| chr7_+_100450328 | 0.19 |
ENST00000540482.1
ENST00000418037.1 ENST00000428758.1 ENST00000275729.3 ENST00000415287.1 ENST00000354161.3 ENST00000416675.1 |
SLC12A9
|
solute carrier family 12, member 9 |
| chr5_-_180229791 | 0.18 |
ENST00000504671.1
ENST00000507384.1 |
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr19_+_1000418 | 0.17 |
ENST00000234389.3
|
GRIN3B
|
glutamate receptor, ionotropic, N-methyl-D-aspartate 3B |
| chr15_-_90294523 | 0.17 |
ENST00000300057.4
|
MESP1
|
mesoderm posterior 1 homolog (mouse) |
| chr19_-_42721819 | 0.17 |
ENST00000336034.4
ENST00000598200.1 ENST00000598727.1 ENST00000596251.1 |
DEDD2
|
death effector domain containing 2 |
| chr6_-_72130472 | 0.17 |
ENST00000426635.2
|
LINC00472
|
long intergenic non-protein coding RNA 472 |
| chr15_+_91416092 | 0.16 |
ENST00000559353.1
|
FURIN
|
furin (paired basic amino acid cleaving enzyme) |
| chr15_+_89182156 | 0.16 |
ENST00000379224.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr17_-_8059638 | 0.16 |
ENST00000584202.1
ENST00000354903.5 ENST00000577253.1 |
PER1
|
period circadian clock 1 |
| chr16_-_69385681 | 0.15 |
ENST00000288025.3
|
TMED6
|
transmembrane emp24 protein transport domain containing 6 |
| chr16_+_30675654 | 0.15 |
ENST00000287468.5
ENST00000395073.2 |
FBRS
|
fibrosin |
| chr19_-_3500635 | 0.15 |
ENST00000250937.3
|
DOHH
|
deoxyhypusine hydroxylase/monooxygenase |
| chrX_-_49056635 | 0.15 |
ENST00000472598.1
ENST00000538567.1 ENST00000479808.1 ENST00000263233.4 |
SYP
|
synaptophysin |
| chr3_-_61237050 | 0.14 |
ENST00000476844.1
ENST00000488467.1 ENST00000492590.1 ENST00000468189.1 |
FHIT
|
fragile histidine triad |
| chr17_-_27230035 | 0.14 |
ENST00000378895.4
ENST00000394901.3 |
DHRS13
|
dehydrogenase/reductase (SDR family) member 13 |
| chr4_-_177116772 | 0.14 |
ENST00000280191.2
|
SPATA4
|
spermatogenesis associated 4 |
| chr17_-_73937028 | 0.14 |
ENST00000586631.2
|
FBF1
|
Fas (TNFRSF6) binding factor 1 |
| chr17_-_72968837 | 0.14 |
ENST00000581676.1
|
HID1
|
HID1 domain containing |
| chr19_-_49622348 | 0.14 |
ENST00000408991.2
|
C19orf73
|
chromosome 19 open reading frame 73 |
| chr12_+_109569155 | 0.14 |
ENST00000539864.1
|
ACACB
|
acetyl-CoA carboxylase beta |
| chr3_+_49726932 | 0.14 |
ENST00000327697.6
ENST00000432042.1 ENST00000454491.1 |
RNF123
|
ring finger protein 123 |
| chr14_+_24779376 | 0.14 |
ENST00000530080.1
|
LTB4R2
|
leukotriene B4 receptor 2 |
| chr19_-_47734448 | 0.14 |
ENST00000439096.2
|
BBC3
|
BCL2 binding component 3 |
| chr16_-_67190152 | 0.13 |
ENST00000486556.1
|
TRADD
|
TNFRSF1A-associated via death domain |
| chr2_-_27486951 | 0.13 |
ENST00000432351.1
|
SLC30A3
|
solute carrier family 30 (zinc transporter), member 3 |
| chr15_+_40733387 | 0.13 |
ENST00000416165.1
|
BAHD1
|
bromo adjacent homology domain containing 1 |
| chr22_-_39268192 | 0.13 |
ENST00000216083.6
|
CBX6
|
chromobox homolog 6 |
| chrX_-_80377162 | 0.13 |
ENST00000430960.1
ENST00000447319.1 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr17_-_7297833 | 0.13 |
ENST00000571802.1
ENST00000576201.1 ENST00000573213.1 ENST00000324822.11 |
TMEM256-PLSCR3
|
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr8_+_87111059 | 0.13 |
ENST00000285393.3
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr19_+_46531127 | 0.13 |
ENST00000601033.1
|
CTC-344H19.4
|
CTC-344H19.4 |
| chrX_-_80377118 | 0.12 |
ENST00000373250.3
|
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr3_-_52719912 | 0.12 |
ENST00000420148.1
|
PBRM1
|
polybromo 1 |
| chr6_-_30043539 | 0.12 |
ENST00000376751.3
ENST00000244360.6 |
RNF39
|
ring finger protein 39 |
| chr20_+_306177 | 0.12 |
ENST00000544632.1
|
SOX12
|
SRY (sex determining region Y)-box 12 |
| chr14_+_24779340 | 0.12 |
ENST00000533293.1
ENST00000543919.1 |
LTB4R2
|
leukotriene B4 receptor 2 |
| chr19_+_18723660 | 0.12 |
ENST00000262817.3
|
TMEM59L
|
transmembrane protein 59-like |
| chr20_-_22565101 | 0.12 |
ENST00000419308.2
|
FOXA2
|
forkhead box A2 |
| chr22_-_39268308 | 0.11 |
ENST00000407418.3
|
CBX6
|
chromobox homolog 6 |
| chr17_+_68164752 | 0.11 |
ENST00000535240.1
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr19_-_49222956 | 0.11 |
ENST00000599703.1
ENST00000318083.6 ENST00000419611.1 ENST00000377367.3 |
MAMSTR
|
MEF2 activating motif and SAP domain containing transcriptional regulator |
| chr17_+_79859985 | 0.11 |
ENST00000333383.7
|
NPB
|
neuropeptide B |
| chr2_+_149402009 | 0.11 |
ENST00000457184.1
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr16_-_11350036 | 0.11 |
ENST00000332029.2
|
SOCS1
|
suppressor of cytokine signaling 1 |
| chr11_-_45939565 | 0.10 |
ENST00000525192.1
ENST00000378750.5 |
PEX16
|
peroxisomal biogenesis factor 16 |
| chr18_+_3450161 | 0.10 |
ENST00000551402.1
ENST00000577543.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr2_+_42275153 | 0.10 |
ENST00000294964.5
|
PKDCC
|
protein kinase domain containing, cytoplasmic |
| chr10_-_102279586 | 0.10 |
ENST00000370345.3
ENST00000451524.1 ENST00000370329.5 |
SEC31B
|
SEC31 homolog B (S. cerevisiae) |
| chr22_-_36013368 | 0.10 |
ENST00000442617.1
ENST00000397326.2 ENST00000397328.1 ENST00000451685.1 |
MB
|
myoglobin |
| chr7_-_55583740 | 0.10 |
ENST00000453256.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr3_-_49726486 | 0.10 |
ENST00000449682.2
|
MST1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr2_-_176046391 | 0.10 |
ENST00000392541.3
ENST00000409194.1 |
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr5_-_180229833 | 0.09 |
ENST00000307826.4
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr16_+_3162557 | 0.09 |
ENST00000382192.3
ENST00000219091.4 ENST00000444510.2 ENST00000414351.1 |
ZNF205
|
zinc finger protein 205 |
| chr12_+_52203789 | 0.09 |
ENST00000599343.1
|
AC068987.1
|
HCG1997999; cDNA FLJ33996 fis, clone DFNES2008881 |
| chr2_-_216003127 | 0.09 |
ENST00000412081.1
ENST00000272895.7 |
ABCA12
|
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr1_-_153521714 | 0.09 |
ENST00000368713.3
|
S100A3
|
S100 calcium binding protein A3 |
| chr16_+_2820912 | 0.09 |
ENST00000570539.1
|
SRRM2
|
serine/arginine repetitive matrix 2 |
| chr20_+_48884002 | 0.09 |
ENST00000425497.1
ENST00000445003.1 |
RP11-290F20.3
|
RP11-290F20.3 |
| chr1_-_154934200 | 0.09 |
ENST00000368457.2
|
PYGO2
|
pygopus family PHD finger 2 |
| chr3_+_52279737 | 0.09 |
ENST00000457351.2
|
PPM1M
|
protein phosphatase, Mg2+/Mn2+ dependent, 1M |
| chr11_+_120081475 | 0.09 |
ENST00000328965.4
|
OAF
|
OAF homolog (Drosophila) |
| chr14_+_85994943 | 0.09 |
ENST00000553678.1
|
RP11-497E19.2
|
Uncharacterized protein |
| chr7_+_2687173 | 0.09 |
ENST00000403167.1
|
TTYH3
|
tweety family member 3 |
| chr1_+_154975110 | 0.08 |
ENST00000535420.1
ENST00000368426.3 |
ZBTB7B
|
zinc finger and BTB domain containing 7B |
| chr16_-_2031464 | 0.08 |
ENST00000356120.4
ENST00000354249.4 |
NOXO1
|
NADPH oxidase organizer 1 |
| chr2_+_171673417 | 0.08 |
ENST00000344257.5
|
GAD1
|
glutamate decarboxylase 1 (brain, 67kDa) |
| chr18_-_60985914 | 0.08 |
ENST00000589955.1
|
BCL2
|
B-cell CLL/lymphoma 2 |
| chr20_+_306221 | 0.08 |
ENST00000342665.2
|
SOX12
|
SRY (sex determining region Y)-box 12 |
| chr1_-_149982624 | 0.08 |
ENST00000417191.1
ENST00000369135.4 |
OTUD7B
|
OTU domain containing 7B |
| chr1_-_153518270 | 0.08 |
ENST00000354332.4
ENST00000368716.4 |
S100A4
|
S100 calcium binding protein A4 |
| chr5_-_148758839 | 0.08 |
ENST00000261796.3
|
IL17B
|
interleukin 17B |
| chr11_+_73358594 | 0.08 |
ENST00000227214.6
ENST00000398494.4 ENST00000543085.1 |
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr17_+_58755184 | 0.08 |
ENST00000589222.1
ENST00000407086.3 ENST00000390652.5 |
BCAS3
|
breast carcinoma amplified sequence 3 |
| chr1_+_8378140 | 0.08 |
ENST00000377479.2
|
SLC45A1
|
solute carrier family 45, member 1 |
| chr19_-_39421377 | 0.08 |
ENST00000430193.3
ENST00000600042.1 ENST00000221431.6 |
SARS2
|
seryl-tRNA synthetase 2, mitochondrial |
| chr12_-_116714564 | 0.08 |
ENST00000548743.1
|
MED13L
|
mediator complex subunit 13-like |
| chr3_-_48632593 | 0.08 |
ENST00000454817.1
ENST00000328333.8 |
COL7A1
|
collagen, type VII, alpha 1 |
| chr17_+_55183261 | 0.08 |
ENST00000576295.1
|
AKAP1
|
A kinase (PRKA) anchor protein 1 |
| chr11_-_45939374 | 0.08 |
ENST00000533151.1
ENST00000241041.3 |
PEX16
|
peroxisomal biogenesis factor 16 |
| chr17_-_15165825 | 0.08 |
ENST00000426385.3
|
PMP22
|
peripheral myelin protein 22 |
| chr1_-_21620877 | 0.08 |
ENST00000527991.1
|
ECE1
|
endothelin converting enzyme 1 |
| chr17_+_77020224 | 0.08 |
ENST00000339142.2
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr8_-_21988558 | 0.08 |
ENST00000312841.8
|
HR
|
hair growth associated |
| chr5_+_32712363 | 0.08 |
ENST00000507141.1
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr12_+_53645870 | 0.07 |
ENST00000329548.4
|
MFSD5
|
major facilitator superfamily domain containing 5 |
| chr3_+_184056614 | 0.07 |
ENST00000453072.1
|
FAM131A
|
family with sequence similarity 131, member A |
| chr1_-_44482979 | 0.07 |
ENST00000360584.2
ENST00000357730.2 ENST00000528803.1 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr3_-_52719810 | 0.07 |
ENST00000424867.1
ENST00000394830.3 ENST00000431678.1 ENST00000450271.1 |
PBRM1
|
polybromo 1 |
| chr22_-_20231207 | 0.07 |
ENST00000425986.1
|
RTN4R
|
reticulon 4 receptor |
| chr17_-_1029866 | 0.07 |
ENST00000570525.1
|
ABR
|
active BCR-related |
| chr4_-_140222358 | 0.07 |
ENST00000505036.1
ENST00000544855.1 ENST00000539002.1 |
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr19_-_8579030 | 0.07 |
ENST00000255616.8
ENST00000393927.4 |
ZNF414
|
zinc finger protein 414 |
| chr2_+_171673072 | 0.07 |
ENST00000358196.3
ENST00000375272.1 |
GAD1
|
glutamate decarboxylase 1 (brain, 67kDa) |
| chr4_-_4249924 | 0.07 |
ENST00000254742.2
ENST00000382753.4 ENST00000540397.1 ENST00000538516.1 |
TMEM128
|
transmembrane protein 128 |
| chr11_+_114310164 | 0.07 |
ENST00000544196.1
ENST00000539754.1 ENST00000539275.1 |
REXO2
|
RNA exonuclease 2 |
| chrX_-_148713365 | 0.07 |
ENST00000511776.1
ENST00000507237.1 |
TMEM185A
|
transmembrane protein 185A |
| chr7_-_92146729 | 0.07 |
ENST00000541751.1
|
PEX1
|
peroxisomal biogenesis factor 1 |
| chr18_+_54318893 | 0.07 |
ENST00000593058.1
|
WDR7
|
WD repeat domain 7 |
| chr1_+_55464600 | 0.07 |
ENST00000371265.4
|
BSND
|
Bartter syndrome, infantile, with sensorineural deafness (Barttin) |
| chr11_-_74660065 | 0.07 |
ENST00000525407.1
ENST00000528219.1 ENST00000531852.1 |
XRRA1
|
X-ray radiation resistance associated 1 |
| chr12_+_57853918 | 0.07 |
ENST00000532291.1
ENST00000543426.1 ENST00000228682.2 ENST00000546141.1 |
GLI1
|
GLI family zinc finger 1 |
| chr22_-_30722912 | 0.07 |
ENST00000215790.7
|
TBC1D10A
|
TBC1 domain family, member 10A |
| chr21_-_46962379 | 0.07 |
ENST00000311124.4
ENST00000380010.4 |
SLC19A1
|
solute carrier family 19 (folate transporter), member 1 |
| chr16_+_57023406 | 0.07 |
ENST00000262510.6
ENST00000308149.7 ENST00000436936.1 |
NLRC5
|
NLR family, CARD domain containing 5 |
| chr17_-_27229948 | 0.07 |
ENST00000426464.2
|
DHRS13
|
dehydrogenase/reductase (SDR family) member 13 |
| chr1_-_151119087 | 0.07 |
ENST00000341697.3
ENST00000368914.3 |
SEMA6C
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6C |
| chr17_+_77020146 | 0.07 |
ENST00000579760.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr1_+_47901689 | 0.07 |
ENST00000334793.5
|
FOXD2
|
forkhead box D2 |
| chr11_+_61129827 | 0.07 |
ENST00000542946.1
|
TMEM138
|
transmembrane protein 138 |
| chr19_+_41107249 | 0.06 |
ENST00000396819.3
|
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chr1_+_1981890 | 0.06 |
ENST00000378567.3
ENST00000468310.1 |
PRKCZ
|
protein kinase C, zeta |
| chr3_-_50360192 | 0.06 |
ENST00000442581.1
ENST00000447092.1 ENST00000357750.4 |
HYAL2
|
hyaluronoglucosaminidase 2 |
| chr4_+_102268904 | 0.06 |
ENST00000527564.1
ENST00000529296.1 |
AP001816.1
|
Uncharacterized protein |
| chr19_+_48867652 | 0.06 |
ENST00000344846.2
|
SYNGR4
|
synaptogyrin 4 |
| chr17_-_7232585 | 0.06 |
ENST00000571887.1
ENST00000315614.7 ENST00000399464.2 ENST00000570460.1 |
NEURL4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr7_+_143318020 | 0.06 |
ENST00000444908.2
ENST00000518791.1 ENST00000411497.2 |
FAM115C
|
family with sequence similarity 115, member C |
| chr1_+_228395755 | 0.06 |
ENST00000284548.11
ENST00000570156.2 ENST00000422127.1 ENST00000366707.4 ENST00000366709.4 |
OBSCN
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| chr4_+_183065793 | 0.06 |
ENST00000512480.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr13_-_28545276 | 0.06 |
ENST00000381020.7
|
CDX2
|
caudal type homeobox 2 |
| chr16_-_2205352 | 0.06 |
ENST00000563192.1
|
RP11-304L19.5
|
RP11-304L19.5 |
| chr20_+_34020827 | 0.06 |
ENST00000374375.1
|
GDF5OS
|
growth differentiation factor 5 opposite strand |
| chr7_+_40174565 | 0.06 |
ENST00000309930.5
ENST00000401647.2 ENST00000335693.4 ENST00000413931.1 ENST00000416370.1 ENST00000540834.1 |
C7orf10
|
succinylCoA:glutarate-CoA transferase |
| chrX_-_153775426 | 0.06 |
ENST00000393562.2
|
G6PD
|
glucose-6-phosphate dehydrogenase |
| chr16_-_70285797 | 0.06 |
ENST00000435634.1
|
EXOSC6
|
exosome component 6 |
| chr2_+_29336855 | 0.06 |
ENST00000404424.1
|
CLIP4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr8_-_145060593 | 0.05 |
ENST00000313059.5
ENST00000524918.1 ENST00000313028.7 ENST00000525773.1 |
PARP10
|
poly (ADP-ribose) polymerase family, member 10 |
| chr17_+_44668387 | 0.05 |
ENST00000576040.1
|
NSF
|
N-ethylmaleimide-sensitive factor |
| chr6_+_150464155 | 0.05 |
ENST00000361131.4
|
PPP1R14C
|
protein phosphatase 1, regulatory (inhibitor) subunit 14C |
| chr6_-_31619697 | 0.05 |
ENST00000434444.1
|
BAG6
|
BCL2-associated athanogene 6 |
| chrX_-_83757399 | 0.05 |
ENST00000373177.2
ENST00000297977.5 ENST00000506585.2 ENST00000449553.2 |
HDX
|
highly divergent homeobox |
| chr2_-_169769787 | 0.05 |
ENST00000451987.1
|
SPC25
|
SPC25, NDC80 kinetochore complex component |
| chr1_-_204329013 | 0.05 |
ENST00000272203.3
ENST00000414478.1 |
PLEKHA6
|
pleckstrin homology domain containing, family A member 6 |
| chr20_+_48909240 | 0.05 |
ENST00000371639.3
|
RP11-290F20.1
|
RP11-290F20.1 |
| chr1_-_6269448 | 0.05 |
ENST00000465335.1
|
RPL22
|
ribosomal protein L22 |
| chr22_-_38539185 | 0.05 |
ENST00000452542.1
|
PLA2G6
|
phospholipase A2, group VI (cytosolic, calcium-independent) |
| chr1_+_154975258 | 0.05 |
ENST00000417934.2
|
ZBTB7B
|
zinc finger and BTB domain containing 7B |
| chr6_-_31619892 | 0.05 |
ENST00000454165.1
ENST00000428326.1 ENST00000452994.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr5_+_82767284 | 0.05 |
ENST00000265077.3
|
VCAN
|
versican |
| chrX_+_102862834 | 0.05 |
ENST00000372627.5
ENST00000243286.3 |
TCEAL3
|
transcription elongation factor A (SII)-like 3 |
| chr9_+_4662282 | 0.05 |
ENST00000381883.2
|
PPAPDC2
|
phosphatidic acid phosphatase type 2 domain containing 2 |
| chr14_-_21994337 | 0.05 |
ENST00000537235.1
ENST00000450879.2 |
SALL2
|
spalt-like transcription factor 2 |
| chr1_-_85930246 | 0.05 |
ENST00000426972.3
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr19_-_10213335 | 0.05 |
ENST00000592641.1
ENST00000253109.4 |
ANGPTL6
|
angiopoietin-like 6 |
| chr17_+_37821593 | 0.05 |
ENST00000578283.1
|
TCAP
|
titin-cap |
| chr17_-_73937116 | 0.05 |
ENST00000586717.1
ENST00000389570.4 ENST00000319129.5 |
FBF1
|
Fas (TNFRSF6) binding factor 1 |
| chrX_+_149887090 | 0.05 |
ENST00000538506.1
|
MTMR1
|
myotubularin related protein 1 |
| chr1_-_15735925 | 0.05 |
ENST00000427824.1
|
RP3-467K16.4
|
RP3-467K16.4 |
| chr6_+_34204642 | 0.05 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr1_+_153963227 | 0.05 |
ENST00000368567.4
ENST00000392558.4 |
RPS27
|
ribosomal protein S27 |
| chr19_-_16045220 | 0.05 |
ENST00000326742.8
|
CYP4F11
|
cytochrome P450, family 4, subfamily F, polypeptide 11 |
| chr17_+_77020325 | 0.05 |
ENST00000311661.4
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr12_-_49351303 | 0.05 |
ENST00000256682.4
|
ARF3
|
ADP-ribosylation factor 3 |
| chr5_+_82767583 | 0.05 |
ENST00000512590.2
ENST00000513960.1 ENST00000513984.1 ENST00000502527.2 |
VCAN
|
versican |
| chr20_-_3185279 | 0.05 |
ENST00000354488.3
ENST00000380201.2 |
DDRGK1
|
DDRGK domain containing 1 |
| chr19_-_42931567 | 0.05 |
ENST00000244289.4
|
LIPE
|
lipase, hormone-sensitive |
| chr19_-_14016877 | 0.05 |
ENST00000454313.1
ENST00000591586.1 ENST00000346736.2 |
C19orf57
|
chromosome 19 open reading frame 57 |
| chr10_+_104263743 | 0.05 |
ENST00000369902.3
ENST00000369899.2 ENST00000423559.2 |
SUFU
|
suppressor of fused homolog (Drosophila) |
| chr2_+_201994042 | 0.05 |
ENST00000417748.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr2_+_11696464 | 0.05 |
ENST00000234142.5
|
GREB1
|
growth regulation by estrogen in breast cancer 1 |
| chr2_+_85646054 | 0.05 |
ENST00000389938.2
|
SH2D6
|
SH2 domain containing 6 |
| chr1_-_157014865 | 0.05 |
ENST00000361409.2
|
ARHGEF11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr14_-_24911868 | 0.04 |
ENST00000554698.1
|
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr11_+_7559485 | 0.04 |
ENST00000527790.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr6_-_130536774 | 0.04 |
ENST00000532763.1
|
SAMD3
|
sterile alpha motif domain containing 3 |
| chr5_-_473135 | 0.04 |
ENST00000342584.3
|
CTD-2228K2.5
|
Uncharacterized protein |
| chr17_-_46703826 | 0.04 |
ENST00000550387.1
ENST00000311177.5 |
HOXB9
|
homeobox B9 |
| chr5_-_176836577 | 0.04 |
ENST00000253496.3
|
F12
|
coagulation factor XII (Hageman factor) |
| chr1_+_155911480 | 0.04 |
ENST00000368318.3
|
RXFP4
|
relaxin/insulin-like family peptide receptor 4 |
| chr12_-_28124903 | 0.04 |
ENST00000395872.1
ENST00000354417.3 ENST00000201015.4 |
PTHLH
|
parathyroid hormone-like hormone |
| chr12_+_57849048 | 0.04 |
ENST00000266646.2
|
INHBE
|
inhibin, beta E |
| chr20_-_30310336 | 0.04 |
ENST00000434194.1
ENST00000376062.2 |
BCL2L1
|
BCL2-like 1 |
| chr19_+_16308711 | 0.04 |
ENST00000429941.2
ENST00000444449.2 ENST00000589822.1 |
AP1M1
|
adaptor-related protein complex 1, mu 1 subunit |
| chrX_-_106960285 | 0.04 |
ENST00000503515.1
ENST00000372397.2 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr7_+_76139833 | 0.04 |
ENST00000257632.5
|
UPK3B
|
uroplakin 3B |
| chrX_-_128788914 | 0.04 |
ENST00000429967.1
ENST00000307484.6 |
APLN
|
apelin |
| chr22_-_46373004 | 0.04 |
ENST00000339464.4
|
WNT7B
|
wingless-type MMTV integration site family, member 7B |
| chr5_-_142784888 | 0.04 |
ENST00000514699.1
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr17_+_57970469 | 0.04 |
ENST00000443572.2
ENST00000406116.3 ENST00000225577.4 ENST00000393021.3 |
RPS6KB1
|
ribosomal protein S6 kinase, 70kDa, polypeptide 1 |
| chr16_+_2587998 | 0.04 |
ENST00000441549.3
ENST00000268673.7 |
PDPK1
|
3-phosphoinositide dependent protein kinase-1 |
| chr8_-_144654918 | 0.04 |
ENST00000529971.1
|
MROH6
|
maestro heat-like repeat family member 6 |
| chr12_+_1738363 | 0.04 |
ENST00000397196.2
|
WNT5B
|
wingless-type MMTV integration site family, member 5B |
| chr13_-_79177673 | 0.04 |
ENST00000377208.5
|
POU4F1
|
POU class 4 homeobox 1 |
| chr1_+_156254070 | 0.04 |
ENST00000405535.2
ENST00000456810.1 |
TMEM79
|
transmembrane protein 79 |
| chr1_+_168250194 | 0.04 |
ENST00000367821.3
|
TBX19
|
T-box 19 |
| chr1_-_157015162 | 0.04 |
ENST00000368194.3
|
ARHGEF11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr3_+_135741576 | 0.04 |
ENST00000334546.2
|
PPP2R3A
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr12_+_56511943 | 0.04 |
ENST00000257940.2
ENST00000552345.1 ENST00000551880.1 ENST00000546903.1 ENST00000551790.1 |
ZC3H10
ESYT1
|
zinc finger CCCH-type containing 10 extended synaptotagmin-like protein 1 |
| chr7_-_105319536 | 0.04 |
ENST00000477775.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr21_+_17214724 | 0.04 |
ENST00000449491.1
|
USP25
|
ubiquitin specific peptidase 25 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX5
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.1 | 0.2 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.1 | 0.2 | GO:0090381 | negative regulation of mesodermal cell fate specification(GO:0042662) cardiac cell fate determination(GO:0060913) regulation of heart induction(GO:0090381) regulation of heart induction by canonical Wnt signaling pathway(GO:0100012) negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.2 | GO:0019082 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.0 | 0.2 | GO:1905205 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) positive regulation of connective tissue replacement(GO:1905205) |
| 0.0 | 0.3 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.2 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.2 | GO:0018352 | glutamate decarboxylation to succinate(GO:0006540) protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.0 | 0.1 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.3 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.3 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.1 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.0 | 0.1 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.2 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.1 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.0 | 0.1 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.1 | GO:0046671 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.0 | 0.2 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.1 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 0.1 | GO:0098838 | reduced folate transmembrane transport(GO:0098838) |
| 0.0 | 0.0 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.1 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.0 | 0.0 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.2 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.0 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.0 | GO:0043396 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.0 | 0.0 | GO:0048633 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 | 0.1 | GO:1904379 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.1 | GO:0010814 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 | 0.1 | GO:0043353 | slow-twitch skeletal muscle fiber contraction(GO:0031444) enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 0.1 | GO:0036309 | protein localization to M-band(GO:0036309) |
| 0.0 | 0.2 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.0 | GO:0042377 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.1 | GO:0010533 | regulation of activation of Janus kinase activity(GO:0010533) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.0 | GO:1903719 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.0 | 0.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.0 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.0 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.1 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.1 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.1 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.0 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) inner medullary collecting duct development(GO:0072061) |
| 0.0 | 0.1 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.1 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.0 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.0 | 0.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.1 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.0 | 0.1 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.1 | 0.3 | GO:0001632 | leukotriene B4 receptor activity(GO:0001632) leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0015633 | zinc transporting ATPase activity(GO:0015633) |
| 0.0 | 0.3 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.3 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.1 | GO:0035034 | histone acetyltransferase regulator activity(GO:0035034) |
| 0.0 | 0.1 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 0.1 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.1 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.0 | 0.1 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.1 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.0 | 0.1 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.0 | 0.1 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.0 | 0.2 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.0 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.3 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.0 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.0 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.0 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |