Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
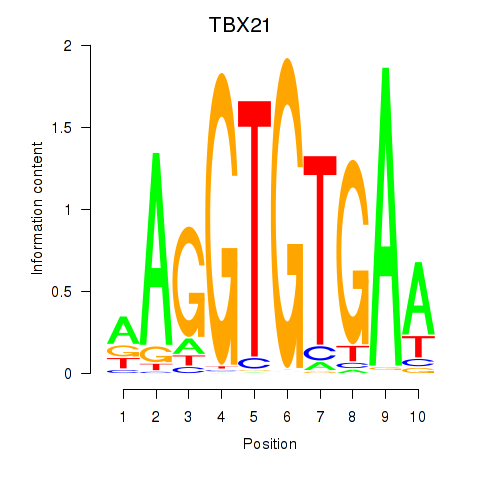
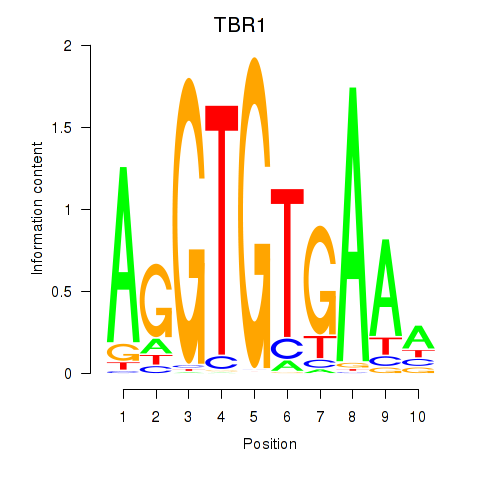
Results for TBX21_TBR1
Z-value: 0.29
Transcription factors associated with TBX21_TBR1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX21
|
ENSG00000073861.2 | T-box transcription factor 21 |
|
TBR1
|
ENSG00000136535.10 | T-box brain transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBR1 | hg19_v2_chr2_+_162272605_162272753 | -0.41 | 5.9e-01 | Click! |
Activity profile of TBX21_TBR1 motif
Sorted Z-values of TBX21_TBR1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_64073699 | 0.30 |
ENST00000405666.1
ENST00000468670.1 |
ESRRA
|
estrogen-related receptor alpha |
| chr15_-_90294523 | 0.28 |
ENST00000300057.4
|
MESP1
|
mesoderm posterior 1 homolog (mouse) |
| chr16_-_67450325 | 0.23 |
ENST00000348579.2
|
ZDHHC1
|
zinc finger, DHHC-type containing 1 |
| chr19_+_36236491 | 0.20 |
ENST00000591949.1
|
PSENEN
|
presenilin enhancer gamma secretase subunit |
| chr15_+_90319557 | 0.20 |
ENST00000341735.3
|
MESP2
|
mesoderm posterior 2 homolog (mouse) |
| chr19_-_7990991 | 0.18 |
ENST00000318978.4
|
CTXN1
|
cortexin 1 |
| chrX_+_47050798 | 0.17 |
ENST00000412206.1
ENST00000427561.1 |
UBA1
|
ubiquitin-like modifier activating enzyme 1 |
| chr15_+_76030311 | 0.15 |
ENST00000543887.1
|
AC019294.1
|
AC019294.1 |
| chr12_+_14572070 | 0.14 |
ENST00000545769.1
ENST00000428217.2 ENST00000396279.2 ENST00000542514.1 ENST00000536279.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr17_+_25799008 | 0.13 |
ENST00000583370.1
ENST00000398988.3 ENST00000268763.6 |
KSR1
|
kinase suppressor of ras 1 |
| chr19_+_56653064 | 0.12 |
ENST00000593100.1
|
ZNF444
|
zinc finger protein 444 |
| chr1_+_156105878 | 0.12 |
ENST00000508500.1
|
LMNA
|
lamin A/C |
| chr16_+_12059091 | 0.12 |
ENST00000562385.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr8_+_144766617 | 0.11 |
ENST00000454097.1
ENST00000534303.1 ENST00000529833.1 ENST00000530574.1 ENST00000442058.2 ENST00000358656.4 ENST00000526970.1 ENST00000532158.1 |
ZNF707
|
zinc finger protein 707 |
| chr16_+_69139467 | 0.11 |
ENST00000569188.1
|
HAS3
|
hyaluronan synthase 3 |
| chr11_-_114271139 | 0.11 |
ENST00000325636.4
|
C11orf71
|
chromosome 11 open reading frame 71 |
| chr8_+_22428457 | 0.11 |
ENST00000517962.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr3_-_66551351 | 0.11 |
ENST00000273261.3
|
LRIG1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr1_-_44482979 | 0.11 |
ENST00000360584.2
ENST00000357730.2 ENST00000528803.1 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr19_-_45748628 | 0.11 |
ENST00000590022.1
|
AC006126.4
|
AC006126.4 |
| chr11_+_120081475 | 0.11 |
ENST00000328965.4
|
OAF
|
OAF homolog (Drosophila) |
| chr9_-_117267717 | 0.11 |
ENST00000374057.3
|
DFNB31
|
deafness, autosomal recessive 31 |
| chr19_+_36236514 | 0.10 |
ENST00000222266.2
|
PSENEN
|
presenilin enhancer gamma secretase subunit |
| chr17_-_7232585 | 0.10 |
ENST00000571887.1
ENST00000315614.7 ENST00000399464.2 ENST00000570460.1 |
NEURL4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr11_-_236326 | 0.10 |
ENST00000525237.1
ENST00000532956.1 ENST00000525319.1 ENST00000524564.1 ENST00000382743.4 |
SIRT3
|
sirtuin 3 |
| chr11_-_60010556 | 0.10 |
ENST00000427611.2
|
MS4A4E
|
membrane-spanning 4-domains, subfamily A, member 4E |
| chr17_-_46657473 | 0.10 |
ENST00000332503.5
|
HOXB4
|
homeobox B4 |
| chr6_+_32132360 | 0.10 |
ENST00000333845.6
ENST00000395512.1 ENST00000432129.1 |
EGFL8
|
EGF-like-domain, multiple 8 |
| chr11_+_61447845 | 0.10 |
ENST00000257215.5
|
DAGLA
|
diacylglycerol lipase, alpha |
| chr1_-_108743471 | 0.09 |
ENST00000569674.1
|
SLC25A24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr17_-_76356148 | 0.09 |
ENST00000587578.1
ENST00000330871.2 |
SOCS3
|
suppressor of cytokine signaling 3 |
| chr1_-_44818599 | 0.09 |
ENST00000537474.1
|
ERI3
|
ERI1 exoribonuclease family member 3 |
| chr16_-_88717482 | 0.09 |
ENST00000261623.3
|
CYBA
|
cytochrome b-245, alpha polypeptide |
| chr11_+_64950801 | 0.09 |
ENST00000526468.1
|
CAPN1
|
calpain 1, (mu/I) large subunit |
| chr3_-_61237050 | 0.09 |
ENST00000476844.1
ENST00000488467.1 ENST00000492590.1 ENST00000468189.1 |
FHIT
|
fragile histidine triad |
| chr19_+_50529329 | 0.09 |
ENST00000599155.1
|
ZNF473
|
zinc finger protein 473 |
| chr22_+_35776354 | 0.08 |
ENST00000412893.1
|
HMOX1
|
heme oxygenase (decycling) 1 |
| chr1_+_28586006 | 0.08 |
ENST00000253063.3
|
SESN2
|
sestrin 2 |
| chr19_+_859654 | 0.08 |
ENST00000592860.1
|
CFD
|
complement factor D (adipsin) |
| chr16_+_30212378 | 0.08 |
ENST00000569485.1
|
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr19_-_3500635 | 0.08 |
ENST00000250937.3
|
DOHH
|
deoxyhypusine hydroxylase/monooxygenase |
| chr17_+_46184911 | 0.08 |
ENST00000580219.1
ENST00000452859.2 ENST00000393405.2 ENST00000439357.2 ENST00000359238.2 |
SNX11
|
sorting nexin 11 |
| chr3_-_66551397 | 0.07 |
ENST00000383703.3
|
LRIG1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr7_+_73106926 | 0.07 |
ENST00000453316.1
|
WBSCR22
|
Williams Beuren syndrome chromosome region 22 |
| chr13_-_30160925 | 0.07 |
ENST00000450494.1
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr11_+_67195917 | 0.07 |
ENST00000524934.1
ENST00000539188.1 ENST00000312629.5 |
RPS6KB2
|
ribosomal protein S6 kinase, 70kDa, polypeptide 2 |
| chr1_+_154975110 | 0.07 |
ENST00000535420.1
ENST00000368426.3 |
ZBTB7B
|
zinc finger and BTB domain containing 7B |
| chr7_+_76101379 | 0.07 |
ENST00000429179.1
|
DTX2
|
deltex homolog 2 (Drosophila) |
| chr11_-_133826852 | 0.07 |
ENST00000533871.2
ENST00000321016.8 |
IGSF9B
|
immunoglobulin superfamily, member 9B |
| chr17_-_43502987 | 0.07 |
ENST00000376922.2
|
ARHGAP27
|
Rho GTPase activating protein 27 |
| chr1_-_85930246 | 0.07 |
ENST00000426972.3
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr11_+_66059339 | 0.07 |
ENST00000327259.4
|
TMEM151A
|
transmembrane protein 151A |
| chr2_+_217498105 | 0.07 |
ENST00000233809.4
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa |
| chr22_-_31688431 | 0.07 |
ENST00000402249.3
ENST00000443175.1 ENST00000215912.5 ENST00000441972.1 |
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr11_-_64052111 | 0.07 |
ENST00000394532.3
ENST00000394531.3 ENST00000309032.3 |
BAD
|
BCL2-associated agonist of cell death |
| chr19_+_38779778 | 0.07 |
ENST00000590738.1
ENST00000587519.2 ENST00000591889.1 |
SPINT2
CTB-102L5.4
|
serine peptidase inhibitor, Kunitz type, 2 CTB-102L5.4 |
| chr11_-_44972476 | 0.07 |
ENST00000527685.1
ENST00000308212.5 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr7_+_140372953 | 0.07 |
ENST00000072869.4
ENST00000476491.1 |
ADCK2
|
aarF domain containing kinase 2 |
| chr8_-_101724989 | 0.06 |
ENST00000517403.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr19_+_6373715 | 0.06 |
ENST00000599849.1
|
ALKBH7
|
alkB, alkylation repair homolog 7 (E. coli) |
| chr15_+_59908633 | 0.06 |
ENST00000559626.1
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr22_+_18560743 | 0.06 |
ENST00000399744.3
|
PEX26
|
peroxisomal biogenesis factor 26 |
| chr19_+_1000418 | 0.06 |
ENST00000234389.3
|
GRIN3B
|
glutamate receptor, ionotropic, N-methyl-D-aspartate 3B |
| chr11_+_64052294 | 0.06 |
ENST00000536667.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr16_+_77233294 | 0.06 |
ENST00000378644.4
|
SYCE1L
|
synaptonemal complex central element protein 1-like |
| chr14_+_95078714 | 0.06 |
ENST00000393078.3
ENST00000393080.4 ENST00000467132.1 |
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 |
| chr11_-_111649074 | 0.06 |
ENST00000534218.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr2_-_74648702 | 0.06 |
ENST00000518863.1
|
C2orf81
|
chromosome 2 open reading frame 81 |
| chr16_-_30546275 | 0.06 |
ENST00000568028.1
|
ZNF747
|
zinc finger protein 747 |
| chrX_+_47077680 | 0.06 |
ENST00000522883.1
|
CDK16
|
cyclin-dependent kinase 16 |
| chr11_+_64052266 | 0.06 |
ENST00000539851.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr19_-_44258770 | 0.06 |
ENST00000601925.1
ENST00000602222.1 ENST00000599804.1 |
SMG9
|
SMG9 nonsense mediated mRNA decay factor |
| chr8_+_22857048 | 0.06 |
ENST00000251822.6
|
RHOBTB2
|
Rho-related BTB domain containing 2 |
| chr22_-_41215328 | 0.06 |
ENST00000434185.1
ENST00000435456.2 |
SLC25A17
|
solute carrier family 25 (mitochondrial carrier; peroxisomal membrane protein, 34kDa), member 17 |
| chr17_+_4046964 | 0.06 |
ENST00000573984.1
|
CYB5D2
|
cytochrome b5 domain containing 2 |
| chr16_-_88717423 | 0.06 |
ENST00000568278.1
ENST00000569359.1 ENST00000567174.1 |
CYBA
|
cytochrome b-245, alpha polypeptide |
| chr12_+_52431016 | 0.06 |
ENST00000553200.1
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr8_+_144766669 | 0.06 |
ENST00000526315.1
ENST00000532205.1 |
ZNF707
|
zinc finger protein 707 |
| chr22_+_35776828 | 0.05 |
ENST00000216117.8
|
HMOX1
|
heme oxygenase (decycling) 1 |
| chr1_-_26633067 | 0.05 |
ENST00000421827.2
ENST00000374215.1 ENST00000374223.1 ENST00000357089.4 ENST00000535108.1 ENST00000314675.7 ENST00000436301.2 ENST00000423664.1 ENST00000374221.3 |
UBXN11
|
UBX domain protein 11 |
| chr3_-_49466686 | 0.05 |
ENST00000273598.3
ENST00000436744.2 |
NICN1
|
nicolin 1 |
| chr11_-_117698765 | 0.05 |
ENST00000532119.1
|
FXYD2
|
FXYD domain containing ion transport regulator 2 |
| chr16_+_50313426 | 0.05 |
ENST00000569265.1
|
ADCY7
|
adenylate cyclase 7 |
| chr11_+_64052454 | 0.05 |
ENST00000539833.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr9_+_35829208 | 0.05 |
ENST00000439587.2
ENST00000377991.4 |
TMEM8B
|
transmembrane protein 8B |
| chr1_+_160313165 | 0.05 |
ENST00000421914.1
ENST00000535857.1 ENST00000438008.1 |
NCSTN
|
nicastrin |
| chr17_-_17726907 | 0.05 |
ENST00000423161.3
|
SREBF1
|
sterol regulatory element binding transcription factor 1 |
| chr7_+_150076406 | 0.05 |
ENST00000329630.5
|
ZNF775
|
zinc finger protein 775 |
| chr10_+_135050908 | 0.05 |
ENST00000325980.9
|
VENTX
|
VENT homeobox |
| chr19_-_10213335 | 0.05 |
ENST00000592641.1
ENST00000253109.4 |
ANGPTL6
|
angiopoietin-like 6 |
| chr19_+_34887220 | 0.05 |
ENST00000592740.1
|
RP11-618P17.4
|
Uncharacterized protein |
| chr1_+_70687137 | 0.05 |
ENST00000436161.2
|
SRSF11
|
serine/arginine-rich splicing factor 11 |
| chr20_+_34680595 | 0.05 |
ENST00000406771.2
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr15_-_41047421 | 0.05 |
ENST00000560460.1
ENST00000338376.3 ENST00000560905.1 |
RMDN3
|
regulator of microtubule dynamics 3 |
| chr2_+_176972000 | 0.05 |
ENST00000249504.5
|
HOXD11
|
homeobox D11 |
| chr12_-_49351303 | 0.05 |
ENST00000256682.4
|
ARF3
|
ADP-ribosylation factor 3 |
| chr22_+_42196666 | 0.05 |
ENST00000402061.3
ENST00000255784.5 |
CCDC134
|
coiled-coil domain containing 134 |
| chr2_+_102758210 | 0.05 |
ENST00000450319.1
|
IL1R1
|
interleukin 1 receptor, type I |
| chr1_-_205744205 | 0.05 |
ENST00000446390.2
|
RAB7L1
|
RAB7, member RAS oncogene family-like 1 |
| chr19_+_48949030 | 0.05 |
ENST00000253237.5
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chr17_-_1553346 | 0.05 |
ENST00000301336.6
|
RILP
|
Rab interacting lysosomal protein |
| chr12_+_57853918 | 0.05 |
ENST00000532291.1
ENST00000543426.1 ENST00000228682.2 ENST00000546141.1 |
GLI1
|
GLI family zinc finger 1 |
| chr2_-_39664206 | 0.05 |
ENST00000484274.1
|
MAP4K3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chr12_+_58148842 | 0.05 |
ENST00000266643.5
|
MARCH9
|
membrane-associated ring finger (C3HC4) 9 |
| chr17_+_8191815 | 0.05 |
ENST00000226105.6
ENST00000407006.4 ENST00000580434.1 ENST00000439238.3 |
RANGRF
|
RAN guanine nucleotide release factor |
| chr2_+_120687335 | 0.05 |
ENST00000544261.1
|
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr17_+_76356516 | 0.05 |
ENST00000592569.1
|
RP11-806H10.4
|
RP11-806H10.4 |
| chr19_-_44258733 | 0.05 |
ENST00000597586.1
ENST00000596714.1 |
SMG9
|
SMG9 nonsense mediated mRNA decay factor |
| chr1_+_20915409 | 0.05 |
ENST00000375071.3
|
CDA
|
cytidine deaminase |
| chr17_+_7358889 | 0.05 |
ENST00000575379.1
|
CHRNB1
|
cholinergic receptor, nicotinic, beta 1 (muscle) |
| chr7_-_87849340 | 0.05 |
ENST00000419179.1
ENST00000265729.2 |
SRI
|
sorcin |
| chr16_+_69221028 | 0.05 |
ENST00000336278.4
|
SNTB2
|
syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2) |
| chr8_-_99955042 | 0.05 |
ENST00000519420.1
|
STK3
|
serine/threonine kinase 3 |
| chr17_-_43209862 | 0.05 |
ENST00000322765.5
|
PLCD3
|
phospholipase C, delta 3 |
| chr6_-_154751629 | 0.04 |
ENST00000424998.1
|
CNKSR3
|
CNKSR family member 3 |
| chr16_-_11350036 | 0.04 |
ENST00000332029.2
|
SOCS1
|
suppressor of cytokine signaling 1 |
| chr2_-_228497888 | 0.04 |
ENST00000264387.4
ENST00000409066.1 |
C2orf83
|
chromosome 2 open reading frame 83 |
| chrX_+_99899180 | 0.04 |
ENST00000373004.3
|
SRPX2
|
sushi-repeat containing protein, X-linked 2 |
| chr17_+_76422409 | 0.04 |
ENST00000600087.1
|
AC061992.1
|
Uncharacterized protein |
| chr11_+_66824303 | 0.04 |
ENST00000533360.1
|
RHOD
|
ras homolog family member D |
| chr9_+_139553306 | 0.04 |
ENST00000371699.1
|
EGFL7
|
EGF-like-domain, multiple 7 |
| chr1_+_144989309 | 0.04 |
ENST00000596396.1
|
AL590452.1
|
Uncharacterized protein |
| chr15_+_76016293 | 0.04 |
ENST00000332145.2
|
ODF3L1
|
outer dense fiber of sperm tails 3-like 1 |
| chr10_-_126432821 | 0.04 |
ENST00000280780.6
|
FAM53B
|
family with sequence similarity 53, member B |
| chr3_+_50654821 | 0.04 |
ENST00000457064.1
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr19_-_35068562 | 0.04 |
ENST00000595013.1
|
SCGB1B2P
|
secretoglobin, family 1B, member 2, pseudogene |
| chr16_-_66968055 | 0.04 |
ENST00000568572.1
|
FAM96B
|
family with sequence similarity 96, member B |
| chr6_-_13487825 | 0.04 |
ENST00000603223.1
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1 |
| chr4_+_108746282 | 0.04 |
ENST00000503862.1
|
SGMS2
|
sphingomyelin synthase 2 |
| chr12_+_65672702 | 0.04 |
ENST00000538045.1
ENST00000535239.1 |
MSRB3
|
methionine sulfoxide reductase B3 |
| chr3_+_50606901 | 0.04 |
ENST00000455834.1
|
HEMK1
|
HemK methyltransferase family member 1 |
| chr11_+_66824346 | 0.04 |
ENST00000532559.1
|
RHOD
|
ras homolog family member D |
| chr11_-_119599794 | 0.04 |
ENST00000264025.3
|
PVRL1
|
poliovirus receptor-related 1 (herpesvirus entry mediator C) |
| chr22_-_38699003 | 0.04 |
ENST00000451964.1
|
CSNK1E
|
casein kinase 1, epsilon |
| chr17_-_38721711 | 0.04 |
ENST00000578085.1
ENST00000246657.2 |
CCR7
|
chemokine (C-C motif) receptor 7 |
| chr9_-_136203235 | 0.04 |
ENST00000372022.4
|
SURF6
|
surfeit 6 |
| chr19_-_10614386 | 0.04 |
ENST00000171111.5
|
KEAP1
|
kelch-like ECH-associated protein 1 |
| chr11_+_66824276 | 0.04 |
ENST00000308831.2
|
RHOD
|
ras homolog family member D |
| chr16_-_30546141 | 0.04 |
ENST00000535210.1
ENST00000395094.3 |
ZNF747
|
zinc finger protein 747 |
| chr3_+_160117062 | 0.04 |
ENST00000497311.1
|
SMC4
|
structural maintenance of chromosomes 4 |
| chr1_-_27701307 | 0.04 |
ENST00000270879.4
ENST00000354982.2 |
FCN3
|
ficolin (collagen/fibrinogen domain containing) 3 |
| chr15_-_52030293 | 0.04 |
ENST00000560491.1
ENST00000267838.3 |
LYSMD2
|
LysM, putative peptidoglycan-binding, domain containing 2 |
| chrX_-_128788914 | 0.04 |
ENST00000429967.1
ENST00000307484.6 |
APLN
|
apelin |
| chr1_+_147013182 | 0.04 |
ENST00000234739.3
|
BCL9
|
B-cell CLL/lymphoma 9 |
| chr9_+_34990219 | 0.04 |
ENST00000541010.1
ENST00000454002.2 ENST00000545841.1 |
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chrX_+_109245863 | 0.04 |
ENST00000372072.3
|
TMEM164
|
transmembrane protein 164 |
| chr19_-_55549624 | 0.04 |
ENST00000417454.1
ENST00000310373.3 ENST00000333884.2 |
GP6
|
glycoprotein VI (platelet) |
| chr17_+_26662679 | 0.04 |
ENST00000578158.1
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr16_+_75690093 | 0.04 |
ENST00000564671.2
|
TERF2IP
|
telomeric repeat binding factor 2, interacting protein |
| chr20_+_44509857 | 0.04 |
ENST00000372523.1
ENST00000372520.1 |
ZSWIM1
|
zinc finger, SWIM-type containing 1 |
| chr11_-_66112555 | 0.04 |
ENST00000425825.2
ENST00000359957.3 |
BRMS1
|
breast cancer metastasis suppressor 1 |
| chr4_-_4249924 | 0.04 |
ENST00000254742.2
ENST00000382753.4 ENST00000540397.1 ENST00000538516.1 |
TMEM128
|
transmembrane protein 128 |
| chr5_+_150639360 | 0.04 |
ENST00000523004.1
|
GM2A
|
GM2 ganglioside activator |
| chr3_-_139195350 | 0.04 |
ENST00000232217.2
|
RBP2
|
retinol binding protein 2, cellular |
| chr11_-_117698787 | 0.04 |
ENST00000260287.2
|
FXYD2
|
FXYD domain containing ion transport regulator 2 |
| chr7_-_20826504 | 0.04 |
ENST00000418710.2
ENST00000361443.4 |
SP8
|
Sp8 transcription factor |
| chr9_+_134378289 | 0.03 |
ENST00000423007.1
ENST00000404875.2 ENST00000441334.1 ENST00000341012.7 ENST00000372228.3 ENST00000402686.3 ENST00000419118.2 ENST00000541219.1 ENST00000354713.4 ENST00000418774.1 ENST00000415075.1 ENST00000448212.1 ENST00000430619.1 |
POMT1
|
protein-O-mannosyltransferase 1 |
| chr1_+_202172848 | 0.03 |
ENST00000255432.7
|
LGR6
|
leucine-rich repeat containing G protein-coupled receptor 6 |
| chr17_+_46185111 | 0.03 |
ENST00000582104.1
ENST00000584335.1 |
SNX11
|
sorting nexin 11 |
| chr11_-_111649015 | 0.03 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr11_+_66790816 | 0.03 |
ENST00000527043.1
|
SYT12
|
synaptotagmin XII |
| chr17_-_37557846 | 0.03 |
ENST00000394294.3
ENST00000583610.1 ENST00000264658.6 |
FBXL20
|
F-box and leucine-rich repeat protein 20 |
| chr9_-_117150303 | 0.03 |
ENST00000312033.3
|
AKNA
|
AT-hook transcription factor |
| chr12_-_76879852 | 0.03 |
ENST00000548341.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr19_+_859425 | 0.03 |
ENST00000327726.6
|
CFD
|
complement factor D (adipsin) |
| chr6_+_26383318 | 0.03 |
ENST00000469230.1
ENST00000490025.1 ENST00000356709.4 ENST00000352867.2 ENST00000493275.1 ENST00000472507.1 ENST00000482536.1 ENST00000432533.2 ENST00000482842.1 |
BTN2A2
|
butyrophilin, subfamily 2, member A2 |
| chr22_+_18560675 | 0.03 |
ENST00000329627.7
|
PEX26
|
peroxisomal biogenesis factor 26 |
| chr1_-_19229014 | 0.03 |
ENST00000538839.1
ENST00000290597.5 |
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr3_+_50606577 | 0.03 |
ENST00000434410.1
ENST00000232854.4 |
HEMK1
|
HemK methyltransferase family member 1 |
| chr16_+_31085714 | 0.03 |
ENST00000300850.5
ENST00000564189.1 ENST00000428260.1 |
ZNF646
|
zinc finger protein 646 |
| chr11_+_45907177 | 0.03 |
ENST00000241014.2
|
MAPK8IP1
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chr12_+_22778291 | 0.03 |
ENST00000545979.1
|
ETNK1
|
ethanolamine kinase 1 |
| chr7_+_100271446 | 0.03 |
ENST00000419828.1
ENST00000427895.1 |
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr1_-_19229248 | 0.03 |
ENST00000375341.3
|
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr3_+_42190714 | 0.03 |
ENST00000449246.1
|
TRAK1
|
trafficking protein, kinesin binding 1 |
| chr13_+_109248500 | 0.03 |
ENST00000356711.2
|
MYO16
|
myosin XVI |
| chr6_+_30848829 | 0.03 |
ENST00000508317.1
|
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr1_-_76076793 | 0.03 |
ENST00000370859.3
|
SLC44A5
|
solute carrier family 44, member 5 |
| chr1_-_19229218 | 0.03 |
ENST00000432718.1
|
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr6_+_43149903 | 0.03 |
ENST00000252050.4
ENST00000354495.3 ENST00000372647.2 |
CUL9
|
cullin 9 |
| chr6_+_30848771 | 0.03 |
ENST00000503180.1
|
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr12_-_58026426 | 0.03 |
ENST00000418555.2
|
B4GALNT1
|
beta-1,4-N-acetyl-galactosaminyl transferase 1 |
| chr1_+_151584544 | 0.03 |
ENST00000458013.2
ENST00000368843.3 |
SNX27
|
sorting nexin family member 27 |
| chr16_-_1429627 | 0.03 |
ENST00000248104.7
|
UNKL
|
unkempt family zinc finger-like |
| chr19_+_10381769 | 0.03 |
ENST00000423829.2
ENST00000588645.1 |
ICAM1
|
intercellular adhesion molecule 1 |
| chr6_+_292253 | 0.03 |
ENST00000603453.1
ENST00000605315.1 ENST00000603881.1 |
DUSP22
|
dual specificity phosphatase 22 |
| chr1_+_33219592 | 0.03 |
ENST00000373481.3
|
KIAA1522
|
KIAA1522 |
| chr11_-_62995986 | 0.03 |
ENST00000403374.2
|
SLC22A25
|
solute carrier family 22, member 25 |
| chr16_+_69140122 | 0.03 |
ENST00000219322.3
|
HAS3
|
hyaluronan synthase 3 |
| chr11_-_6440624 | 0.03 |
ENST00000311051.3
|
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr11_-_1783633 | 0.03 |
ENST00000367196.3
|
CTSD
|
cathepsin D |
| chr9_-_215744 | 0.03 |
ENST00000382387.2
|
C9orf66
|
chromosome 9 open reading frame 66 |
| chr5_-_119669160 | 0.03 |
ENST00000514240.1
|
CTC-552D5.1
|
CTC-552D5.1 |
| chr7_+_102290772 | 0.03 |
ENST00000507450.1
|
SPDYE2B
|
speedy/RINGO cell cycle regulator family member E2B |
| chr17_-_79358929 | 0.03 |
ENST00000570301.1
|
RP11-1055B8.3
|
RP11-1055B8.3 |
| chr19_+_3762703 | 0.03 |
ENST00000589174.1
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chrX_-_154376044 | 0.03 |
ENST00000362018.2
|
MTCP1
|
mature T-cell proliferation 1 |
| chr3_-_50360165 | 0.03 |
ENST00000428028.1
|
HYAL2
|
hyaluronoglucosaminidase 2 |
| chr1_-_36020531 | 0.03 |
ENST00000440579.1
ENST00000494948.1 |
KIAA0319L
|
KIAA0319-like |
| chr11_-_33913708 | 0.03 |
ENST00000257818.2
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX21_TBR1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0090381 | negative regulation of mesodermal cell fate specification(GO:0042662) cardiac cell fate determination(GO:0060913) regulation of heart induction(GO:0090381) regulation of heart induction by canonical Wnt signaling pathway(GO:0100012) negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.2 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.0 | 0.1 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.2 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.1 | GO:0006788 | heme oxidation(GO:0006788) negative regulation of mast cell cytokine production(GO:0032764) regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.1 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.1 | GO:0043132 | coenzyme A transport(GO:0015880) coenzyme A transmembrane transport(GO:0035349) NAD transport(GO:0043132) adenosine 3',5'-bisphosphate transmembrane transport(GO:0071106) AMP transport(GO:0080121) |
| 0.0 | 0.1 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.1 | GO:0060139 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.0 | 0.0 | GO:0006337 | nucleosome disassembly(GO:0006337) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.1 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.0 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.0 | 0.1 | GO:0006562 | proline catabolic process(GO:0006562) proline catabolic process to glutamate(GO:0010133) 4-hydroxyproline catabolic process(GO:0019470) |
| 0.0 | 0.0 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.4 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.0 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.1 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.0 | GO:0051466 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.0 | 0.1 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.0 | 0.1 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.0 | GO:0070839 | divalent metal ion export(GO:0070839) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.1 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.0 | 0.0 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.0 | 0.4 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.1 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.1 | GO:0003842 | 1-pyrroline-5-carboxylate dehydrogenase activity(GO:0003842) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.1 | GO:0071077 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.0 | 0.1 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.0 | 0.1 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.1 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0033906 | protein tyrosine kinase inhibitor activity(GO:0030292) hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |