Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
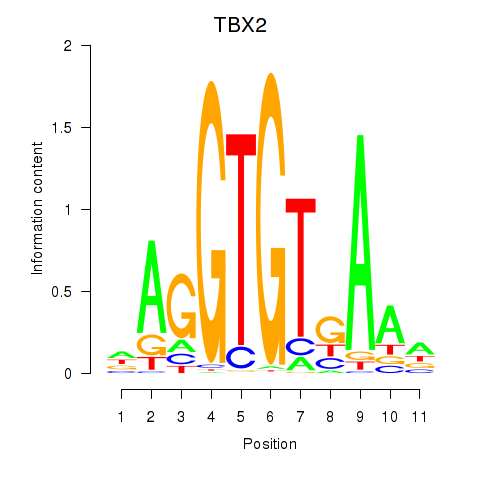
Results for TBX2
Z-value: 0.55
Transcription factors associated with TBX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX2
|
ENSG00000121068.9 | T-box transcription factor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBX2 | hg19_v2_chr17_+_59477233_59477263 | -0.81 | 1.9e-01 | Click! |
Activity profile of TBX2 motif
Sorted Z-values of TBX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_29033682 | 0.35 |
ENST00000379579.4
ENST00000334056.5 ENST00000449210.1 |
SPDYA
|
speedy/RINGO cell cycle regulator family member A |
| chr19_-_51875894 | 0.22 |
ENST00000600427.1
ENST00000595217.1 ENST00000221978.5 |
NKG7
|
natural killer cell group 7 sequence |
| chr7_+_107220899 | 0.21 |
ENST00000379117.2
ENST00000473124.1 |
BCAP29
|
B-cell receptor-associated protein 29 |
| chr3_-_194207388 | 0.20 |
ENST00000457986.1
|
ATP13A3
|
ATPase type 13A3 |
| chr5_+_81601166 | 0.20 |
ENST00000439350.1
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like |
| chr1_+_182758900 | 0.17 |
ENST00000367555.1
ENST00000367554.3 |
NPL
|
N-acetylneuraminate pyruvate lyase (dihydrodipicolinate synthase) |
| chr9_+_26956474 | 0.17 |
ENST00000429045.2
|
IFT74
|
intraflagellar transport 74 homolog (Chlamydomonas) |
| chr14_+_21467414 | 0.17 |
ENST00000554422.1
ENST00000298681.4 |
SLC39A2
|
solute carrier family 39 (zinc transporter), member 2 |
| chr12_-_107380910 | 0.16 |
ENST00000392830.2
ENST00000240050.4 |
MTERFD3
|
MTERF domain containing 3 |
| chr10_+_94833642 | 0.16 |
ENST00000224356.4
ENST00000394139.1 |
CYP26A1
|
cytochrome P450, family 26, subfamily A, polypeptide 1 |
| chr8_-_99954788 | 0.16 |
ENST00000523601.1
|
STK3
|
serine/threonine kinase 3 |
| chr1_+_35734616 | 0.15 |
ENST00000441447.1
|
ZMYM4
|
zinc finger, MYM-type 4 |
| chr19_-_43702231 | 0.15 |
ENST00000597374.1
ENST00000599371.1 |
PSG4
|
pregnancy specific beta-1-glycoprotein 4 |
| chr10_-_14596140 | 0.15 |
ENST00000496330.1
|
FAM107B
|
family with sequence similarity 107, member B |
| chrX_-_46187069 | 0.15 |
ENST00000446884.1
|
RP1-30G7.2
|
RP1-30G7.2 |
| chr4_-_89619386 | 0.15 |
ENST00000323061.5
|
NAP1L5
|
nucleosome assembly protein 1-like 5 |
| chr17_+_39868577 | 0.14 |
ENST00000329402.3
|
GAST
|
gastrin |
| chr1_+_212782012 | 0.14 |
ENST00000341491.4
ENST00000366985.1 |
ATF3
|
activating transcription factor 3 |
| chr11_-_74204692 | 0.14 |
ENST00000528085.1
|
LIPT2
|
lipoyl(octanoyl) transferase 2 (putative) |
| chr6_-_31651817 | 0.14 |
ENST00000375863.3
ENST00000375860.2 |
LY6G5C
|
lymphocyte antigen 6 complex, locus G5C |
| chr1_+_178482262 | 0.14 |
ENST00000367641.3
ENST00000367639.1 |
TEX35
|
testis expressed 35 |
| chr3_+_32726774 | 0.14 |
ENST00000538368.1
|
CNOT10
|
CCR4-NOT transcription complex, subunit 10 |
| chr10_-_112678692 | 0.14 |
ENST00000605742.1
|
BBIP1
|
BBSome interacting protein 1 |
| chr14_+_104177607 | 0.14 |
ENST00000429169.1
|
AL049840.1
|
Uncharacterized protein; cDNA FLJ53535 |
| chrX_-_119693745 | 0.14 |
ENST00000371323.2
|
CUL4B
|
cullin 4B |
| chr8_-_124428569 | 0.13 |
ENST00000521903.1
|
ATAD2
|
ATPase family, AAA domain containing 2 |
| chr13_-_52027134 | 0.13 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr10_-_112678904 | 0.13 |
ENST00000423273.1
ENST00000436562.1 ENST00000447005.1 ENST00000454061.1 |
BBIP1
|
BBSome interacting protein 1 |
| chr21_+_30502806 | 0.13 |
ENST00000399928.1
ENST00000399926.1 |
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr1_+_9599540 | 0.13 |
ENST00000302692.6
|
SLC25A33
|
solute carrier family 25 (pyrimidine nucleotide carrier), member 33 |
| chr6_-_84937314 | 0.13 |
ENST00000257766.4
ENST00000403245.3 |
KIAA1009
|
KIAA1009 |
| chr11_+_111749650 | 0.13 |
ENST00000528125.1
|
C11orf1
|
chromosome 11 open reading frame 1 |
| chr22_-_51001332 | 0.13 |
ENST00000406915.3
|
SYCE3
|
synaptonemal complex central element protein 3 |
| chr10_-_112678976 | 0.12 |
ENST00000448814.1
|
BBIP1
|
BBSome interacting protein 1 |
| chr19_-_44174330 | 0.12 |
ENST00000340093.3
|
PLAUR
|
plasminogen activator, urokinase receptor |
| chr11_+_20409070 | 0.12 |
ENST00000331079.6
|
PRMT3
|
protein arginine methyltransferase 3 |
| chr14_+_77582905 | 0.12 |
ENST00000557408.1
|
TMEM63C
|
transmembrane protein 63C |
| chr9_+_131452239 | 0.12 |
ENST00000372688.4
ENST00000372686.5 |
SET
|
SET nuclear oncogene |
| chr9_+_40028620 | 0.12 |
ENST00000426179.1
|
AL353791.1
|
AL353791.1 |
| chr19_+_19779589 | 0.12 |
ENST00000541458.1
|
ZNF101
|
zinc finger protein 101 |
| chr12_-_46385811 | 0.12 |
ENST00000419565.2
|
SCAF11
|
SR-related CTD-associated factor 11 |
| chr12_+_54379569 | 0.12 |
ENST00000513209.1
|
RP11-834C11.12
|
RP11-834C11.12 |
| chr3_-_119379427 | 0.11 |
ENST00000264231.3
ENST00000468801.1 ENST00000538678.1 |
POPDC2
|
popeye domain containing 2 |
| chr9_+_123837141 | 0.11 |
ENST00000373865.2
|
CNTRL
|
centriolin |
| chr7_-_27135591 | 0.11 |
ENST00000343060.4
ENST00000355633.5 |
HOXA1
|
homeobox A1 |
| chr7_-_76255444 | 0.11 |
ENST00000454397.1
|
POMZP3
|
POM121 and ZP3 fusion |
| chr12_-_12849073 | 0.11 |
ENST00000332427.2
ENST00000540796.1 |
GPR19
|
G protein-coupled receptor 19 |
| chr3_+_160117087 | 0.10 |
ENST00000357388.3
|
SMC4
|
structural maintenance of chromosomes 4 |
| chr4_+_38511367 | 0.10 |
ENST00000507056.1
|
RP11-213G21.1
|
RP11-213G21.1 |
| chr3_+_142315294 | 0.10 |
ENST00000464320.1
|
PLS1
|
plastin 1 |
| chr17_-_8066843 | 0.10 |
ENST00000404970.3
|
VAMP2
|
vesicle-associated membrane protein 2 (synaptobrevin 2) |
| chr19_-_44174305 | 0.10 |
ENST00000601723.1
ENST00000339082.3 |
PLAUR
|
plasminogen activator, urokinase receptor |
| chr12_+_93965609 | 0.10 |
ENST00000549887.1
ENST00000551556.1 |
SOCS2
|
suppressor of cytokine signaling 2 |
| chr3_+_32726620 | 0.10 |
ENST00000331889.6
ENST00000328834.5 |
CNOT10
|
CCR4-NOT transcription complex, subunit 10 |
| chr17_+_25958174 | 0.10 |
ENST00000313648.6
ENST00000577392.1 ENST00000584661.1 ENST00000413914.2 |
LGALS9
|
lectin, galactoside-binding, soluble, 9 |
| chr8_+_82192501 | 0.10 |
ENST00000297258.6
|
FABP5
|
fatty acid binding protein 5 (psoriasis-associated) |
| chr19_-_6670128 | 0.10 |
ENST00000245912.3
|
TNFSF14
|
tumor necrosis factor (ligand) superfamily, member 14 |
| chr2_-_105953912 | 0.10 |
ENST00000610036.1
|
RP11-332H14.2
|
RP11-332H14.2 |
| chr14_-_69619291 | 0.10 |
ENST00000554215.1
ENST00000556847.1 |
DCAF5
|
DDB1 and CUL4 associated factor 5 |
| chr21_+_30503282 | 0.10 |
ENST00000399925.1
|
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr7_+_108210012 | 0.10 |
ENST00000249356.3
|
DNAJB9
|
DnaJ (Hsp40) homolog, subfamily B, member 9 |
| chr15_-_83315874 | 0.10 |
ENST00000569257.1
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr7_+_107220660 | 0.09 |
ENST00000465919.1
ENST00000445771.2 ENST00000479917.1 ENST00000421217.1 ENST00000457837.1 |
BCAP29
|
B-cell receptor-associated protein 29 |
| chr11_+_114271251 | 0.09 |
ENST00000375490.5
|
RBM7
|
RNA binding motif protein 7 |
| chr11_-_92931098 | 0.09 |
ENST00000326402.4
|
SLC36A4
|
solute carrier family 36 (proton/amino acid symporter), member 4 |
| chr22_+_18593097 | 0.09 |
ENST00000426208.1
|
TUBA8
|
tubulin, alpha 8 |
| chr11_-_85779971 | 0.09 |
ENST00000393346.3
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr7_+_110731062 | 0.09 |
ENST00000308478.5
ENST00000451085.1 ENST00000422987.3 ENST00000421101.1 |
LRRN3
|
leucine rich repeat neuronal 3 |
| chr1_+_38022572 | 0.09 |
ENST00000541606.1
|
DNALI1
|
dynein, axonemal, light intermediate chain 1 |
| chr5_-_1551802 | 0.09 |
ENST00000503113.1
|
CTD-2245E15.3
|
CTD-2245E15.3 |
| chr9_+_131447342 | 0.09 |
ENST00000409104.3
|
SET
|
SET nuclear oncogene |
| chr1_+_222988464 | 0.09 |
ENST00000420335.1
|
RP11-452F19.3
|
RP11-452F19.3 |
| chr4_+_124317940 | 0.09 |
ENST00000505319.1
ENST00000339241.1 |
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
| chr1_+_19967014 | 0.09 |
ENST00000428975.1
|
NBL1
|
neuroblastoma 1, DAN family BMP antagonist |
| chr2_-_175869936 | 0.09 |
ENST00000409900.3
|
CHN1
|
chimerin 1 |
| chrX_-_106146547 | 0.09 |
ENST00000276173.4
ENST00000411805.1 |
RIPPLY1
|
ripply transcriptional repressor 1 |
| chr11_+_65266507 | 0.09 |
ENST00000544868.1
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr1_-_159915386 | 0.09 |
ENST00000361509.3
ENST00000368094.1 |
IGSF9
|
immunoglobulin superfamily, member 9 |
| chr7_-_108210048 | 0.09 |
ENST00000415914.3
ENST00000438865.1 |
THAP5
|
THAP domain containing 5 |
| chr19_-_14628645 | 0.09 |
ENST00000598235.1
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr7_-_108209897 | 0.09 |
ENST00000313516.5
|
THAP5
|
THAP domain containing 5 |
| chr7_+_35840819 | 0.08 |
ENST00000399035.3
|
SEPT7
|
septin 7 |
| chr4_+_26578293 | 0.08 |
ENST00000512840.1
|
TBC1D19
|
TBC1 domain family, member 19 |
| chr14_-_45603657 | 0.08 |
ENST00000396062.3
|
FKBP3
|
FK506 binding protein 3, 25kDa |
| chr6_-_105584560 | 0.08 |
ENST00000336775.5
|
BVES
|
blood vessel epicardial substance |
| chr10_+_71444655 | 0.08 |
ENST00000434931.2
|
RP11-242G20.1
|
Uncharacterized protein |
| chr11_+_114270752 | 0.08 |
ENST00000540163.1
|
RBM7
|
RNA binding motif protein 7 |
| chr11_+_55653396 | 0.08 |
ENST00000244891.3
|
TRIM51
|
tripartite motif-containing 51 |
| chr20_-_271009 | 0.08 |
ENST00000382369.5
|
C20orf96
|
chromosome 20 open reading frame 96 |
| chr16_-_18418092 | 0.08 |
ENST00000545050.1
|
NPIPA8
|
nuclear pore complex interacting protein family, member A8 |
| chrX_+_16804544 | 0.08 |
ENST00000380122.5
ENST00000398155.4 |
TXLNG
|
taxilin gamma |
| chr6_-_110500905 | 0.08 |
ENST00000392587.2
|
WASF1
|
WAS protein family, member 1 |
| chr2_+_108905095 | 0.08 |
ENST00000251481.6
ENST00000326853.5 |
SULT1C2
|
sulfotransferase family, cytosolic, 1C, member 2 |
| chr4_-_2243839 | 0.08 |
ENST00000511885.2
ENST00000506763.1 ENST00000514395.1 ENST00000502440.1 ENST00000243706.4 ENST00000443786.2 |
POLN
HAUS3
|
polymerase (DNA directed) nu HAUS augmin-like complex, subunit 3 |
| chr6_-_110500826 | 0.08 |
ENST00000265601.3
ENST00000447287.1 ENST00000444391.1 |
WASF1
|
WAS protein family, member 1 |
| chr10_+_60028818 | 0.08 |
ENST00000333926.5
|
CISD1
|
CDGSH iron sulfur domain 1 |
| chr19_+_56270380 | 0.08 |
ENST00000434937.2
|
RFPL4A
|
ret finger protein-like 4A |
| chr1_-_47779762 | 0.08 |
ENST00000371877.3
ENST00000360380.3 ENST00000337817.5 ENST00000447475.2 |
STIL
|
SCL/TAL1 interrupting locus |
| chr16_-_15463926 | 0.08 |
ENST00000432570.2
|
NPIPA5
|
nuclear pore complex interacting protein family, member A5 |
| chr10_+_95256356 | 0.08 |
ENST00000371485.3
|
CEP55
|
centrosomal protein 55kDa |
| chr4_-_83931862 | 0.08 |
ENST00000506560.1
ENST00000442461.2 ENST00000446851.2 ENST00000340417.3 |
LIN54
|
lin-54 homolog (C. elegans) |
| chr19_-_51875523 | 0.08 |
ENST00000593572.1
ENST00000595157.1 |
NKG7
|
natural killer cell group 7 sequence |
| chr12_+_57854274 | 0.08 |
ENST00000528432.1
|
GLI1
|
GLI family zinc finger 1 |
| chr3_+_142315225 | 0.08 |
ENST00000457734.2
ENST00000483373.1 ENST00000475296.1 ENST00000495744.1 ENST00000476044.1 ENST00000461644.1 |
PLS1
|
plastin 1 |
| chr11_-_16430399 | 0.08 |
ENST00000528252.1
|
SOX6
|
SRY (sex determining region Y)-box 6 |
| chr19_-_44405941 | 0.08 |
ENST00000587128.1
|
RP11-15A1.3
|
RP11-15A1.3 |
| chr10_+_127512106 | 0.08 |
ENST00000278100.6
ENST00000299130.3 ENST00000368759.5 ENST00000429863.2 |
BCCIP
|
BRCA2 and CDKN1A interacting protein |
| chr4_+_155484155 | 0.08 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr11_+_117947724 | 0.07 |
ENST00000534111.1
|
TMPRSS4
|
transmembrane protease, serine 4 |
| chr13_+_98628886 | 0.07 |
ENST00000490680.1
ENST00000539640.1 ENST00000403772.3 |
IPO5
|
importin 5 |
| chr19_+_13049413 | 0.07 |
ENST00000316448.5
ENST00000588454.1 |
CALR
|
calreticulin |
| chr5_-_55290773 | 0.07 |
ENST00000502326.3
ENST00000381298.2 |
IL6ST
|
interleukin 6 signal transducer (gp130, oncostatin M receptor) |
| chr17_+_30771279 | 0.07 |
ENST00000261712.3
ENST00000578213.1 ENST00000457654.2 ENST00000579451.1 |
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr2_-_175870085 | 0.07 |
ENST00000409156.3
|
CHN1
|
chimerin 1 |
| chrX_-_119709637 | 0.07 |
ENST00000404115.3
|
CUL4B
|
cullin 4B |
| chr17_-_1463095 | 0.07 |
ENST00000575895.1
ENST00000573056.1 |
PITPNA
|
phosphatidylinositol transfer protein, alpha |
| chrX_+_18443703 | 0.07 |
ENST00000379996.3
|
CDKL5
|
cyclin-dependent kinase-like 5 |
| chr17_+_16945820 | 0.07 |
ENST00000577514.1
|
MPRIP
|
myosin phosphatase Rho interacting protein |
| chr17_-_60005365 | 0.07 |
ENST00000444766.3
|
INTS2
|
integrator complex subunit 2 |
| chr2_+_228736335 | 0.07 |
ENST00000440997.1
ENST00000545118.1 |
DAW1
|
dynein assembly factor with WDR repeat domains 1 |
| chr9_-_32552551 | 0.07 |
ENST00000360538.2
ENST00000379858.1 |
TOPORS
|
topoisomerase I binding, arginine/serine-rich, E3 ubiquitin protein ligase |
| chr2_+_234875252 | 0.07 |
ENST00000456930.1
|
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr12_+_32112340 | 0.07 |
ENST00000540924.1
ENST00000312561.4 |
KIAA1551
|
KIAA1551 |
| chr9_+_26956371 | 0.07 |
ENST00000380062.5
ENST00000518614.1 |
IFT74
|
intraflagellar transport 74 homolog (Chlamydomonas) |
| chr14_+_90864504 | 0.07 |
ENST00000544280.2
|
CALM1
|
calmodulin 1 (phosphorylase kinase, delta) |
| chr8_+_96037205 | 0.07 |
ENST00000396124.4
|
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr7_+_141438393 | 0.07 |
ENST00000484178.1
ENST00000473783.1 ENST00000481508.1 |
SSBP1
|
single-stranded DNA binding protein 1, mitochondrial |
| chr3_+_121554046 | 0.07 |
ENST00000273668.2
ENST00000451944.2 |
EAF2
|
ELL associated factor 2 |
| chr1_+_35225339 | 0.07 |
ENST00000339480.1
|
GJB4
|
gap junction protein, beta 4, 30.3kDa |
| chr4_+_39408470 | 0.07 |
ENST00000257408.4
|
KLB
|
klotho beta |
| chr1_+_32716857 | 0.07 |
ENST00000482949.1
ENST00000495610.2 |
LCK
|
lymphocyte-specific protein tyrosine kinase |
| chrX_-_55515635 | 0.07 |
ENST00000500968.3
|
USP51
|
ubiquitin specific peptidase 51 |
| chr1_+_35246775 | 0.07 |
ENST00000373366.2
|
GJB3
|
gap junction protein, beta 3, 31kDa |
| chr14_-_35183755 | 0.07 |
ENST00000555765.1
|
CFL2
|
cofilin 2 (muscle) |
| chr4_-_185303418 | 0.07 |
ENST00000610223.1
ENST00000608785.1 |
RP11-290F5.1
|
RP11-290F5.1 |
| chr11_+_108535752 | 0.07 |
ENST00000322536.3
|
DDX10
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 10 |
| chr11_+_122709200 | 0.07 |
ENST00000227348.4
|
CRTAM
|
cytotoxic and regulatory T cell molecule |
| chr14_-_69619689 | 0.07 |
ENST00000389997.6
ENST00000557386.1 ENST00000554681.1 |
DCAF5
|
DDB1 and CUL4 associated factor 5 |
| chr1_+_35734562 | 0.07 |
ENST00000314607.6
ENST00000373297.2 |
ZMYM4
|
zinc finger, MYM-type 4 |
| chr7_-_35840198 | 0.07 |
ENST00000412856.1
ENST00000437235.3 ENST00000424194.1 |
AC007551.3
|
AC007551.3 |
| chr2_-_114461655 | 0.06 |
ENST00000424612.1
|
AC017074.2
|
AC017074.2 |
| chr8_+_1993173 | 0.06 |
ENST00000523438.1
|
MYOM2
|
myomesin 2 |
| chr1_-_211752073 | 0.06 |
ENST00000367001.4
|
SLC30A1
|
solute carrier family 30 (zinc transporter), member 1 |
| chr19_-_39108568 | 0.06 |
ENST00000586296.1
|
MAP4K1
|
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr17_-_7081435 | 0.06 |
ENST00000380920.4
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr4_+_141445311 | 0.06 |
ENST00000323570.3
ENST00000511887.2 |
ELMOD2
|
ELMO/CED-12 domain containing 2 |
| chr1_+_89150245 | 0.06 |
ENST00000370513.5
|
PKN2
|
protein kinase N2 |
| chr6_+_84222220 | 0.06 |
ENST00000369700.3
|
PRSS35
|
protease, serine, 35 |
| chr7_+_99102267 | 0.06 |
ENST00000326775.5
ENST00000451158.1 |
ZKSCAN5
|
zinc finger with KRAB and SCAN domains 5 |
| chr1_+_200993071 | 0.06 |
ENST00000446333.1
ENST00000458003.1 |
RP11-168O16.1
|
RP11-168O16.1 |
| chr3_-_105588231 | 0.06 |
ENST00000545639.1
ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr15_+_41913690 | 0.06 |
ENST00000563576.1
|
MGA
|
MGA, MAX dimerization protein |
| chr17_+_79031415 | 0.06 |
ENST00000572073.1
ENST00000573677.1 |
BAIAP2
|
BAI1-associated protein 2 |
| chr2_-_209118974 | 0.06 |
ENST00000415913.1
ENST00000415282.1 ENST00000446179.1 |
IDH1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr14_-_69619823 | 0.06 |
ENST00000341516.5
|
DCAF5
|
DDB1 and CUL4 associated factor 5 |
| chr1_+_203444887 | 0.06 |
ENST00000343110.2
|
PRELP
|
proline/arginine-rich end leucine-rich repeat protein |
| chr7_+_143013198 | 0.06 |
ENST00000343257.2
|
CLCN1
|
chloride channel, voltage-sensitive 1 |
| chr13_+_73629107 | 0.06 |
ENST00000539231.1
|
KLF5
|
Kruppel-like factor 5 (intestinal) |
| chr14_+_32546274 | 0.06 |
ENST00000396582.2
|
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr15_-_55581954 | 0.06 |
ENST00000336787.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr1_+_32716840 | 0.06 |
ENST00000336890.5
|
LCK
|
lymphocyte-specific protein tyrosine kinase |
| chr1_+_160051319 | 0.06 |
ENST00000368088.3
|
KCNJ9
|
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr7_+_35840542 | 0.06 |
ENST00000435235.1
ENST00000399034.2 ENST00000350320.6 ENST00000469679.2 |
SEPT7
|
septin 7 |
| chr7_-_143892748 | 0.06 |
ENST00000378115.2
|
ARHGEF35
|
Rho guanine nucleotide exchange factor (GEF) 35 |
| chr5_-_145562147 | 0.06 |
ENST00000545646.1
ENST00000274562.9 ENST00000510191.1 ENST00000394434.2 |
LARS
|
leucyl-tRNA synthetase |
| chr2_-_120124258 | 0.06 |
ENST00000409877.1
ENST00000409523.1 ENST00000409466.2 ENST00000414534.1 |
C2orf76
|
chromosome 2 open reading frame 76 |
| chr2_+_189156586 | 0.06 |
ENST00000409830.1
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr1_-_13698405 | 0.06 |
ENST00000540591.1
|
PRAMEF19
|
PRAME family member 19 |
| chr12_-_45270151 | 0.06 |
ENST00000429094.2
|
NELL2
|
NEL-like 2 (chicken) |
| chr3_+_167453493 | 0.06 |
ENST00000295777.5
ENST00000472747.2 |
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr1_+_38022513 | 0.06 |
ENST00000296218.7
|
DNALI1
|
dynein, axonemal, light intermediate chain 1 |
| chr19_-_50529193 | 0.06 |
ENST00000596445.1
ENST00000599538.1 |
VRK3
|
vaccinia related kinase 3 |
| chr13_+_76378305 | 0.06 |
ENST00000526371.1
ENST00000526528.1 |
LMO7
|
LIM domain 7 |
| chr9_+_34329493 | 0.06 |
ENST00000379158.2
ENST00000346365.4 ENST00000379155.5 |
NUDT2
|
nudix (nucleoside diphosphate linked moiety X)-type motif 2 |
| chr1_-_113247543 | 0.06 |
ENST00000414971.1
ENST00000534717.1 |
RHOC
|
ras homolog family member C |
| chr2_-_136743436 | 0.06 |
ENST00000441323.1
ENST00000449218.1 |
DARS
|
aspartyl-tRNA synthetase |
| chr2_-_148778323 | 0.06 |
ENST00000440042.1
ENST00000535373.1 ENST00000540442.1 ENST00000536575.1 |
ORC4
|
origin recognition complex, subunit 4 |
| chr2_+_233320827 | 0.06 |
ENST00000295463.3
|
ALPI
|
alkaline phosphatase, intestinal |
| chr1_+_206858232 | 0.06 |
ENST00000294981.4
|
MAPKAPK2
|
mitogen-activated protein kinase-activated protein kinase 2 |
| chr2_-_61245363 | 0.06 |
ENST00000316752.6
|
PUS10
|
pseudouridylate synthase 10 |
| chr11_-_112034831 | 0.06 |
ENST00000280357.7
|
IL18
|
interleukin 18 (interferon-gamma-inducing factor) |
| chr1_+_201708992 | 0.06 |
ENST00000367295.1
|
NAV1
|
neuron navigator 1 |
| chr19_+_52264449 | 0.05 |
ENST00000599326.1
ENST00000598953.1 |
FPR2
|
formyl peptide receptor 2 |
| chr16_+_640201 | 0.05 |
ENST00000563109.1
|
RAB40C
|
RAB40C, member RAS oncogene family |
| chr2_+_234601512 | 0.05 |
ENST00000305139.6
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr9_+_140032842 | 0.05 |
ENST00000371561.3
ENST00000315048.3 |
GRIN1
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1 |
| chr2_-_39664405 | 0.05 |
ENST00000341681.5
ENST00000263881.3 |
MAP4K3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chr2_-_191878874 | 0.05 |
ENST00000392322.3
ENST00000392323.2 ENST00000424722.1 ENST00000361099.3 |
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr4_+_155484103 | 0.05 |
ENST00000302068.4
|
FGB
|
fibrinogen beta chain |
| chr19_+_50180507 | 0.05 |
ENST00000454376.2
ENST00000524771.1 |
PRMT1
|
protein arginine methyltransferase 1 |
| chr13_+_27825706 | 0.05 |
ENST00000272274.4
ENST00000319826.4 ENST00000326092.4 |
RPL21
|
ribosomal protein L21 |
| chr1_-_200589859 | 0.05 |
ENST00000367350.4
|
KIF14
|
kinesin family member 14 |
| chr1_+_145507587 | 0.05 |
ENST00000330165.8
ENST00000369307.3 |
RBM8A
|
RNA binding motif protein 8A |
| chr12_-_56120838 | 0.05 |
ENST00000548160.1
|
CD63
|
CD63 molecule |
| chr11_-_7847519 | 0.05 |
ENST00000328375.1
|
OR5P3
|
olfactory receptor, family 5, subfamily P, member 3 |
| chr9_-_103115135 | 0.05 |
ENST00000537512.1
|
TEX10
|
testis expressed 10 |
| chr12_+_75874984 | 0.05 |
ENST00000550491.1
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chrX_-_133930285 | 0.05 |
ENST00000486347.1
ENST00000343004.5 |
FAM122B
|
family with sequence similarity 122B |
| chr11_+_107643129 | 0.05 |
ENST00000447610.1
|
AP001024.2
|
Uncharacterized protein |
| chr7_-_99766191 | 0.05 |
ENST00000423751.1
ENST00000360039.4 |
GAL3ST4
|
galactose-3-O-sulfotransferase 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.1 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 0.1 | GO:0002519 | natural killer cell tolerance induction(GO:0002519) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.0 | 0.2 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.0 | 0.2 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.2 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.0 | 0.2 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.3 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.2 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.1 | GO:0002501 | MHC protein complex assembly(GO:0002396) peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.0 | 0.2 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.1 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.0 | 0.1 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.0 | 0.2 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.1 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0006429 | glutaminyl-tRNA aminoacylation(GO:0006425) leucyl-tRNA aminoacylation(GO:0006429) |
| 0.0 | 0.1 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.1 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.1 | GO:0043449 | cellular alkene metabolic process(GO:0043449) |
| 0.0 | 0.1 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.0 | 0.1 | GO:0033504 | floor plate development(GO:0033504) |
| 0.0 | 0.2 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.1 | GO:0002447 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.0 | 0.1 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.1 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.1 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.0 | 0.1 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.0 | 0.1 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.0 | 0.1 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.1 | GO:0061101 | neuroendocrine cell differentiation(GO:0061101) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.0 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.1 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.1 | GO:0061687 | detoxification of inorganic compound(GO:0061687) stress response to metal ion(GO:0097501) |
| 0.0 | 0.1 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.1 | GO:0035720 | intraciliary anterograde transport(GO:0035720) ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.0 | GO:0019364 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.0 | 0.1 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.4 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.1 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.0 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.0 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.0 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 0.2 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.0 | 0.2 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.2 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.2 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.1 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.0 | 0.1 | GO:0004823 | glutamine-tRNA ligase activity(GO:0004819) leucine-tRNA ligase activity(GO:0004823) |
| 0.0 | 0.1 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) leukemia inhibitory factor receptor activity(GO:0004923) interleukin-6 binding(GO:0019981) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.0 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.0 | GO:0008184 | purine nucleobase binding(GO:0002060) glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.1 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.0 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.1 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |