Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
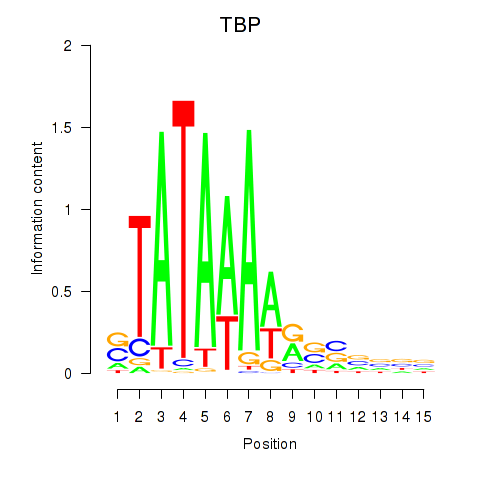
Results for TBP
Z-value: 3.24
Transcription factors associated with TBP
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBP
|
ENSG00000112592.8 | TATA-box binding protein |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBP | hg19_v2_chr6_+_170863672_170863698 | -0.41 | 5.9e-01 | Click! |
Activity profile of TBP motif
Sorted Z-values of TBP motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_149785236 | 2.96 |
ENST00000331491.1
|
HIST2H3D
|
histone cluster 2, H3d |
| chr6_+_26156551 | 2.75 |
ENST00000304218.3
|
HIST1H1E
|
histone cluster 1, H1e |
| chr6_-_26235206 | 2.70 |
ENST00000244534.5
|
HIST1H1D
|
histone cluster 1, H1d |
| chr6_-_26032288 | 2.23 |
ENST00000244661.2
|
HIST1H3B
|
histone cluster 1, H3b |
| chr6_+_27114861 | 2.19 |
ENST00000377459.1
|
HIST1H2AH
|
histone cluster 1, H2ah |
| chr5_+_53686658 | 2.15 |
ENST00000512618.1
|
LINC01033
|
long intergenic non-protein coding RNA 1033 |
| chr6_+_26104104 | 2.06 |
ENST00000377803.2
|
HIST1H4C
|
histone cluster 1, H4c |
| chr6_-_26189304 | 1.98 |
ENST00000340756.2
|
HIST1H4D
|
histone cluster 1, H4d |
| chr6_-_26033796 | 1.96 |
ENST00000259791.2
|
HIST1H2AB
|
histone cluster 1, H2ab |
| chr6_-_27835357 | 1.88 |
ENST00000331442.3
|
HIST1H1B
|
histone cluster 1, H1b |
| chr6_+_26045603 | 1.77 |
ENST00000540144.1
|
HIST1H3C
|
histone cluster 1, H3c |
| chr6_-_26250835 | 1.71 |
ENST00000446824.2
|
HIST1H3F
|
histone cluster 1, H3f |
| chr6_-_26043885 | 1.60 |
ENST00000357905.2
|
HIST1H2BB
|
histone cluster 1, H2bb |
| chr8_-_6735451 | 1.54 |
ENST00000297439.3
|
DEFB1
|
defensin, beta 1 |
| chr6_-_26216872 | 1.52 |
ENST00000244601.3
|
HIST1H2BG
|
histone cluster 1, H2bg |
| chr17_-_34207295 | 1.48 |
ENST00000463941.1
ENST00000293272.3 |
CCL5
|
chemokine (C-C motif) ligand 5 |
| chr18_+_68002675 | 1.44 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chr6_+_26183958 | 1.43 |
ENST00000356530.3
|
HIST1H2BE
|
histone cluster 1, H2be |
| chr7_+_143013198 | 1.40 |
ENST00000343257.2
|
CLCN1
|
chloride channel, voltage-sensitive 1 |
| chr9_-_21077939 | 1.39 |
ENST00000380232.2
|
IFNB1
|
interferon, beta 1, fibroblast |
| chr9_-_35658007 | 1.38 |
ENST00000602361.1
|
RMRP
|
RNA component of mitochondrial RNA processing endoribonuclease |
| chr17_-_39661849 | 1.37 |
ENST00000246635.3
ENST00000336861.3 ENST00000587544.1 ENST00000587435.1 |
KRT13
|
keratin 13 |
| chr6_-_27860956 | 1.37 |
ENST00000359611.2
|
HIST1H2AM
|
histone cluster 1, H2am |
| chr7_+_112120908 | 1.34 |
ENST00000439068.2
ENST00000312849.4 ENST00000429049.1 |
LSMEM1
|
leucine-rich single-pass membrane protein 1 |
| chr11_+_65266507 | 1.27 |
ENST00000544868.1
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr6_-_27100529 | 1.22 |
ENST00000607124.1
ENST00000339812.2 ENST00000541790.1 |
HIST1H2BJ
|
histone cluster 1, H2bj |
| chr9_-_140142222 | 1.17 |
ENST00000344774.4
ENST00000388932.2 |
FAM166A
|
family with sequence similarity 166, member A |
| chr12_+_52695617 | 1.14 |
ENST00000293525.5
|
KRT86
|
keratin 86 |
| chr6_-_26285737 | 1.14 |
ENST00000377727.1
ENST00000289352.1 |
HIST1H4H
|
histone cluster 1, H4h |
| chr17_+_16284749 | 1.13 |
ENST00000578706.1
|
UBB
|
ubiquitin B |
| chr11_-_18270182 | 1.13 |
ENST00000528349.1
ENST00000526900.1 ENST00000529528.1 ENST00000414546.2 ENST00000256733.4 |
SAA2
|
serum amyloid A2 |
| chr14_+_75745477 | 1.13 |
ENST00000303562.4
ENST00000554617.1 ENST00000554212.1 ENST00000535987.1 ENST00000555242.1 |
FOS
|
FBJ murine osteosarcoma viral oncogene homolog |
| chr19_+_35773242 | 1.13 |
ENST00000222304.3
|
HAMP
|
hepcidin antimicrobial peptide |
| chr1_+_33546714 | 1.11 |
ENST00000294517.6
ENST00000358680.3 ENST00000373443.3 ENST00000398167.1 |
ADC
|
arginine decarboxylase |
| chr11_+_18287801 | 1.06 |
ENST00000532858.1
ENST00000405158.2 |
SAA1
|
serum amyloid A1 |
| chr17_-_79792909 | 1.04 |
ENST00000330261.4
ENST00000570394.1 |
PPP1R27
|
protein phosphatase 1, regulatory subunit 27 |
| chr1_-_149812765 | 1.03 |
ENST00000369158.1
|
HIST2H3C
|
histone cluster 2, H3c |
| chr6_+_26021869 | 1.03 |
ENST00000359907.3
|
HIST1H4A
|
histone cluster 1, H4a |
| chr6_-_26124138 | 0.99 |
ENST00000314332.5
ENST00000396984.1 |
HIST1H2BC
|
histone cluster 1, H2bc |
| chr10_+_96698406 | 0.99 |
ENST00000260682.6
|
CYP2C9
|
cytochrome P450, family 2, subfamily C, polypeptide 9 |
| chr17_+_74729060 | 0.97 |
ENST00000587459.1
|
RP11-318A15.7
|
Uncharacterized protein |
| chr6_+_26204825 | 0.96 |
ENST00000360441.4
|
HIST1H4E
|
histone cluster 1, H4e |
| chr11_-_67415048 | 0.94 |
ENST00000529256.1
|
ACY3
|
aspartoacylase (aminocyclase) 3 |
| chr17_+_16284604 | 0.93 |
ENST00000395839.1
ENST00000395837.1 |
UBB
|
ubiquitin B |
| chr17_-_39661947 | 0.93 |
ENST00000590425.1
|
KRT13
|
keratin 13 |
| chr12_-_49582593 | 0.87 |
ENST00000295766.5
|
TUBA1A
|
tubulin, alpha 1a |
| chr1_-_23886285 | 0.87 |
ENST00000374561.5
|
ID3
|
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein |
| chr12_+_11905413 | 0.86 |
ENST00000545027.1
|
ETV6
|
ets variant 6 |
| chr7_-_3214287 | 0.86 |
ENST00000404626.3
|
AC091801.1
|
LOC392621; Uncharacterized protein |
| chr6_+_27100811 | 0.84 |
ENST00000359193.2
|
HIST1H2AG
|
histone cluster 1, H2ag |
| chr11_-_9025541 | 0.83 |
ENST00000525100.1
ENST00000309166.3 ENST00000531090.1 |
NRIP3
|
nuclear receptor interacting protein 3 |
| chr6_-_27840099 | 0.77 |
ENST00000328488.2
|
HIST1H3I
|
histone cluster 1, H3i |
| chr8_+_99956759 | 0.76 |
ENST00000522510.1
ENST00000457907.2 |
OSR2
|
odd-skipped related transciption factor 2 |
| chr1_-_149814478 | 0.76 |
ENST00000369161.3
|
HIST2H2AA3
|
histone cluster 2, H2aa3 |
| chr9_-_21228221 | 0.76 |
ENST00000413767.2
|
IFNA17
|
interferon, alpha 17 |
| chr6_+_27107053 | 0.75 |
ENST00000354348.2
|
HIST1H4I
|
histone cluster 1, H4i |
| chr7_+_116593433 | 0.73 |
ENST00000323984.3
ENST00000393449.1 |
ST7
|
suppression of tumorigenicity 7 |
| chr1_+_149822620 | 0.73 |
ENST00000369159.2
|
HIST2H2AA4
|
histone cluster 2, H2aa4 |
| chr6_+_27833034 | 0.72 |
ENST00000357320.2
|
HIST1H2AL
|
histone cluster 1, H2al |
| chr7_+_116593292 | 0.71 |
ENST00000393446.2
ENST00000265437.5 ENST00000393451.3 |
ST7
|
suppression of tumorigenicity 7 |
| chr6_+_26199737 | 0.71 |
ENST00000359985.1
|
HIST1H2BF
|
histone cluster 1, H2bf |
| chr1_-_207226313 | 0.70 |
ENST00000367084.1
|
YOD1
|
YOD1 deubiquitinase |
| chr5_-_158757895 | 0.69 |
ENST00000231228.2
|
IL12B
|
interleukin 12B (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation factor 2, p40) |
| chrX_+_18725758 | 0.69 |
ENST00000472826.1
ENST00000544635.1 ENST00000496075.2 |
PPEF1
|
protein phosphatase, EF-hand calcium binding domain 1 |
| chr2_-_27531313 | 0.69 |
ENST00000296099.2
|
UCN
|
urocortin |
| chr11_+_18287721 | 0.68 |
ENST00000356524.4
|
SAA1
|
serum amyloid A1 |
| chr10_-_8095412 | 0.67 |
ENST00000458727.1
ENST00000355358.1 |
RP11-379F12.3
GATA3-AS1
|
RP11-379F12.3 GATA3 antisense RNA 1 |
| chr19_+_2476116 | 0.67 |
ENST00000215631.4
ENST00000587345.1 |
GADD45B
|
growth arrest and DNA-damage-inducible, beta |
| chr4_+_74606223 | 0.66 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr6_-_52859968 | 0.65 |
ENST00000370959.1
|
GSTA4
|
glutathione S-transferase alpha 4 |
| chr1_+_202830876 | 0.64 |
ENST00000456105.2
|
RP11-480I12.7
|
RP11-480I12.7 |
| chr1_+_92414952 | 0.63 |
ENST00000449584.1
ENST00000427104.1 ENST00000355011.3 ENST00000448194.1 ENST00000426141.1 ENST00000450792.1 ENST00000548992.1 ENST00000552654.1 ENST00000457265.1 |
BRDT
|
bromodomain, testis-specific |
| chr7_+_115862858 | 0.63 |
ENST00000393481.2
|
TES
|
testis derived transcript (3 LIM domains) |
| chr16_-_67965756 | 0.63 |
ENST00000571044.1
ENST00000571605.1 |
CTRL
|
chymotrypsin-like |
| chr17_+_39421591 | 0.63 |
ENST00000391355.1
|
KRTAP9-6
|
keratin associated protein 9-6 |
| chr1_-_1009683 | 0.62 |
ENST00000453464.2
|
RNF223
|
ring finger protein 223 |
| chr17_-_73150629 | 0.62 |
ENST00000356033.4
ENST00000405458.3 ENST00000409753.3 |
HN1
|
hematological and neurological expressed 1 |
| chr1_-_20755140 | 0.62 |
ENST00000418743.1
ENST00000426428.1 |
RP4-749H3.1
|
long intergenic non-protein coding RNA 1141 |
| chr17_-_73150599 | 0.61 |
ENST00000392566.2
ENST00000581874.1 |
HN1
|
hematological and neurological expressed 1 |
| chr17_+_16284104 | 0.60 |
ENST00000577958.1
ENST00000302182.3 ENST00000577640.1 |
UBB
|
ubiquitin B |
| chr16_-_56701933 | 0.59 |
ENST00000568675.1
ENST00000569500.1 ENST00000444837.2 ENST00000379811.3 |
MT1G
|
metallothionein 1G |
| chr20_+_2276639 | 0.59 |
ENST00000381458.5
|
TGM3
|
transglutaminase 3 |
| chr17_-_38545799 | 0.58 |
ENST00000577541.1
|
TOP2A
|
topoisomerase (DNA) II alpha 170kDa |
| chr6_-_134495992 | 0.56 |
ENST00000475719.2
ENST00000367857.5 ENST00000237305.7 |
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr17_+_39405939 | 0.55 |
ENST00000334109.2
|
KRTAP9-4
|
keratin associated protein 9-4 |
| chr18_-_33077868 | 0.55 |
ENST00000590757.1
ENST00000592173.1 ENST00000441607.2 ENST00000587450.1 ENST00000589258.1 |
INO80C
RP11-322E11.6
|
INO80 complex subunit C Uncharacterized protein |
| chr22_-_40929812 | 0.55 |
ENST00000422851.1
|
MKL1
|
megakaryoblastic leukemia (translocation) 1 |
| chr11_+_62432777 | 0.54 |
ENST00000532971.1
|
METTL12
|
methyltransferase like 12 |
| chr19_+_8386371 | 0.54 |
ENST00000600659.2
|
RPS28
|
ribosomal protein S28 |
| chr11_+_62623621 | 0.54 |
ENST00000535296.1
|
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr8_-_23021533 | 0.53 |
ENST00000312584.3
|
TNFRSF10D
|
tumor necrosis factor receptor superfamily, member 10d, decoy with truncated death domain |
| chr6_-_27806117 | 0.53 |
ENST00000330180.2
|
HIST1H2AK
|
histone cluster 1, H2ak |
| chr12_-_52845910 | 0.52 |
ENST00000252252.3
|
KRT6B
|
keratin 6B |
| chr19_+_13049413 | 0.52 |
ENST00000316448.5
ENST00000588454.1 |
CALR
|
calreticulin |
| chr17_+_79849872 | 0.52 |
ENST00000584197.1
ENST00000583839.1 |
ANAPC11
|
anaphase promoting complex subunit 11 |
| chr1_-_179457805 | 0.51 |
ENST00000600581.1
|
AL160286.1
|
Uncharacterized protein |
| chr6_+_27861190 | 0.50 |
ENST00000303806.4
|
HIST1H2BO
|
histone cluster 1, H2bo |
| chr20_+_30193083 | 0.50 |
ENST00000376112.3
ENST00000376105.3 |
ID1
|
inhibitor of DNA binding 1, dominant negative helix-loop-helix protein |
| chr3_+_193853927 | 0.50 |
ENST00000232424.3
|
HES1
|
hes family bHLH transcription factor 1 |
| chr3_-_170587815 | 0.50 |
ENST00000466674.1
|
RPL22L1
|
ribosomal protein L22-like 1 |
| chr6_-_116833500 | 0.49 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr11_-_36531774 | 0.49 |
ENST00000348124.5
ENST00000526995.1 |
TRAF6
|
TNF receptor-associated factor 6, E3 ubiquitin protein ligase |
| chr5_-_74162739 | 0.49 |
ENST00000513277.1
|
FAM169A
|
family with sequence similarity 169, member A |
| chrX_+_72783026 | 0.48 |
ENST00000373504.6
ENST00000373502.5 |
CHIC1
|
cysteine-rich hydrophobic domain 1 |
| chr3_+_62304712 | 0.48 |
ENST00000494481.1
|
C3orf14
|
chromosome 3 open reading frame 14 |
| chrX_-_55515635 | 0.47 |
ENST00000500968.3
|
USP51
|
ubiquitin specific peptidase 51 |
| chr19_+_36347787 | 0.47 |
ENST00000347900.6
ENST00000360202.5 |
KIRREL2
|
kin of IRRE like 2 (Drosophila) |
| chr19_+_12944722 | 0.47 |
ENST00000591495.1
|
MAST1
|
microtubule associated serine/threonine kinase 1 |
| chr8_-_30002179 | 0.45 |
ENST00000320542.3
|
MBOAT4
|
membrane bound O-acyltransferase domain containing 4 |
| chr11_+_62623544 | 0.45 |
ENST00000377890.2
ENST00000377891.2 ENST00000377889.2 |
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr15_+_76629064 | 0.45 |
ENST00000290759.4
|
ISL2
|
ISL LIM homeobox 2 |
| chr19_+_46367518 | 0.44 |
ENST00000302177.2
|
FOXA3
|
forkhead box A3 |
| chr6_+_30585486 | 0.44 |
ENST00000259873.4
ENST00000506373.2 |
MRPS18B
|
mitochondrial ribosomal protein S18B |
| chr11_+_62623512 | 0.44 |
ENST00000377892.1
|
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr17_+_77021702 | 0.44 |
ENST00000392445.2
ENST00000354124.3 |
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr17_+_79849523 | 0.44 |
ENST00000572639.1
ENST00000579978.1 ENST00000344877.5 ENST00000582222.1 ENST00000577425.1 ENST00000571024.2 ENST00000574924.2 ENST00000572851.2 ENST00000357385.3 ENST00000584314.1 ENST00000571874.2 ENST00000578550.1 ENST00000577747.1 ENST00000579133.1 |
ANAPC11
|
anaphase promoting complex subunit 11 |
| chr8_-_82598067 | 0.43 |
ENST00000523942.1
ENST00000522997.1 |
IMPA1
|
inositol(myo)-1(or 4)-monophosphatase 1 |
| chr6_-_3227877 | 0.43 |
ENST00000259818.7
|
TUBB2B
|
tubulin, beta 2B class IIb |
| chr3_-_170587974 | 0.43 |
ENST00000463836.1
|
RPL22L1
|
ribosomal protein L22-like 1 |
| chr6_+_64282447 | 0.43 |
ENST00000370650.2
ENST00000578299.1 |
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr12_-_21757774 | 0.43 |
ENST00000261195.2
|
GYS2
|
glycogen synthase 2 (liver) |
| chr3_-_172859017 | 0.42 |
ENST00000351008.3
|
SPATA16
|
spermatogenesis associated 16 |
| chr7_+_75931861 | 0.42 |
ENST00000248553.6
|
HSPB1
|
heat shock 27kDa protein 1 |
| chr17_+_16284399 | 0.41 |
ENST00000535788.1
|
UBB
|
ubiquitin B |
| chr12_-_49393092 | 0.41 |
ENST00000421952.2
|
DDN
|
dendrin |
| chr19_-_6720686 | 0.41 |
ENST00000245907.6
|
C3
|
complement component 3 |
| chr11_-_76155700 | 0.41 |
ENST00000572035.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr19_+_1524072 | 0.41 |
ENST00000454744.2
|
PLK5
|
polo-like kinase 5 |
| chr17_+_79849782 | 0.41 |
ENST00000392376.3
|
ANAPC11
|
anaphase promoting complex subunit 11 |
| chr1_-_95007193 | 0.40 |
ENST00000370207.4
ENST00000334047.7 |
F3
|
coagulation factor III (thromboplastin, tissue factor) |
| chr17_+_29248918 | 0.40 |
ENST00000581548.1
ENST00000580525.1 |
ADAP2
|
ArfGAP with dual PH domains 2 |
| chr10_-_101190202 | 0.40 |
ENST00000543866.1
ENST00000370508.5 |
GOT1
|
glutamic-oxaloacetic transaminase 1, soluble |
| chrX_-_7895755 | 0.39 |
ENST00000444736.1
ENST00000537427.1 ENST00000442940.1 |
PNPLA4
|
patatin-like phospholipase domain containing 4 |
| chr19_-_6767516 | 0.39 |
ENST00000245908.6
|
SH2D3A
|
SH2 domain containing 3A |
| chr19_+_50922187 | 0.39 |
ENST00000595883.1
ENST00000597855.1 ENST00000596074.1 ENST00000439922.2 ENST00000594685.1 ENST00000270632.7 |
SPIB
|
Spi-B transcription factor (Spi-1/PU.1 related) |
| chr3_+_70048881 | 0.39 |
ENST00000483525.1
|
RP11-460N16.1
|
RP11-460N16.1 |
| chr1_-_117753540 | 0.38 |
ENST00000328189.3
ENST00000369458.3 |
VTCN1
|
V-set domain containing T cell activation inhibitor 1 |
| chr12_-_49582837 | 0.37 |
ENST00000547939.1
ENST00000546918.1 ENST00000552924.1 |
TUBA1A
|
tubulin, alpha 1a |
| chr11_+_3875757 | 0.37 |
ENST00000525403.1
|
STIM1
|
stromal interaction molecule 1 |
| chr2_-_21266816 | 0.36 |
ENST00000399256.4
|
APOB
|
apolipoprotein B |
| chr8_+_120428546 | 0.36 |
ENST00000259526.3
|
NOV
|
nephroblastoma overexpressed |
| chr6_+_111195973 | 0.36 |
ENST00000368885.3
ENST00000368882.3 ENST00000451850.2 ENST00000368877.5 |
AMD1
|
adenosylmethionine decarboxylase 1 |
| chr1_-_11907829 | 0.36 |
ENST00000376480.3
|
NPPA
|
natriuretic peptide A |
| chr13_+_27844464 | 0.36 |
ENST00000241463.4
|
RASL11A
|
RAS-like, family 11, member A |
| chr1_-_226187013 | 0.36 |
ENST00000272091.7
|
SDE2
|
SDE2 telomere maintenance homolog (S. pombe) |
| chr21_-_28217721 | 0.36 |
ENST00000284984.3
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr1_+_40942887 | 0.35 |
ENST00000372706.1
|
ZFP69
|
ZFP69 zinc finger protein |
| chr8_+_1993173 | 0.35 |
ENST00000523438.1
|
MYOM2
|
myomesin 2 |
| chr4_+_74735102 | 0.35 |
ENST00000395761.3
|
CXCL1
|
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
| chr12_+_13044787 | 0.35 |
ENST00000534831.1
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A |
| chr18_+_616711 | 0.35 |
ENST00000579494.1
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr1_+_86046433 | 0.35 |
ENST00000451137.2
|
CYR61
|
cysteine-rich, angiogenic inducer, 61 |
| chr22_+_19706958 | 0.34 |
ENST00000395109.2
|
SEPT5
|
septin 5 |
| chr9_-_130639997 | 0.34 |
ENST00000373176.1
|
AK1
|
adenylate kinase 1 |
| chr6_+_122720681 | 0.34 |
ENST00000368455.4
ENST00000452194.1 |
HSF2
|
heat shock transcription factor 2 |
| chr17_-_10560619 | 0.34 |
ENST00000583535.1
|
MYH3
|
myosin, heavy chain 3, skeletal muscle, embryonic |
| chr11_-_4719072 | 0.34 |
ENST00000396950.3
ENST00000532598.1 |
OR51E2
|
olfactory receptor, family 51, subfamily E, member 2 |
| chr22_-_19466732 | 0.34 |
ENST00000263202.10
ENST00000360834.4 |
UFD1L
|
ubiquitin fusion degradation 1 like (yeast) |
| chr17_+_7184986 | 0.33 |
ENST00000317370.8
ENST00000571308.1 |
SLC2A4
|
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr20_-_23731893 | 0.33 |
ENST00000398402.1
|
CST1
|
cystatin SN |
| chr1_-_71513471 | 0.33 |
ENST00000370931.3
ENST00000356595.4 ENST00000306666.5 ENST00000370932.2 ENST00000351052.5 ENST00000414819.1 ENST00000370924.4 |
PTGER3
|
prostaglandin E receptor 3 (subtype EP3) |
| chr12_-_118796910 | 0.33 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr21_-_43786634 | 0.33 |
ENST00000291527.2
|
TFF1
|
trefoil factor 1 |
| chr16_+_89989687 | 0.33 |
ENST00000315491.7
ENST00000555576.1 ENST00000554336.1 ENST00000553967.1 |
TUBB3
|
Tubulin beta-3 chain |
| chr6_-_33239712 | 0.32 |
ENST00000436044.2
|
VPS52
|
vacuolar protein sorting 52 homolog (S. cerevisiae) |
| chr4_-_84035905 | 0.32 |
ENST00000311507.4
|
PLAC8
|
placenta-specific 8 |
| chr7_+_72742178 | 0.32 |
ENST00000442793.1
ENST00000413573.2 ENST00000252037.4 |
FKBP6
|
FK506 binding protein 6, 36kDa |
| chr17_-_39538550 | 0.32 |
ENST00000394001.1
|
KRT34
|
keratin 34 |
| chr9_+_135937365 | 0.31 |
ENST00000372080.4
ENST00000351304.7 |
CEL
|
carboxyl ester lipase |
| chr1_+_225140372 | 0.31 |
ENST00000366848.1
ENST00000439375.2 |
DNAH14
|
dynein, axonemal, heavy chain 14 |
| chr1_+_173837488 | 0.30 |
ENST00000427304.1
ENST00000432989.1 ENST00000367702.1 |
ZBTB37
|
zinc finger and BTB domain containing 37 |
| chr17_-_39781054 | 0.30 |
ENST00000463128.1
|
KRT17
|
keratin 17 |
| chr16_+_56691911 | 0.30 |
ENST00000568475.1
|
MT1F
|
metallothionein 1F |
| chr19_+_2269485 | 0.30 |
ENST00000582888.4
ENST00000602676.2 ENST00000322297.4 ENST00000583542.4 |
OAZ1
|
ornithine decarboxylase antizyme 1 |
| chr17_-_48278983 | 0.30 |
ENST00000225964.5
|
COL1A1
|
collagen, type I, alpha 1 |
| chr20_+_18488528 | 0.30 |
ENST00000377465.1
|
SEC23B
|
Sec23 homolog B (S. cerevisiae) |
| chr20_-_43753104 | 0.29 |
ENST00000372785.3
|
WFDC12
|
WAP four-disulfide core domain 12 |
| chr7_-_134143841 | 0.29 |
ENST00000285930.4
|
AKR1B1
|
aldo-keto reductase family 1, member B1 (aldose reductase) |
| chr8_+_1993152 | 0.29 |
ENST00000262113.4
|
MYOM2
|
myomesin 2 |
| chr6_-_33239612 | 0.29 |
ENST00000482399.1
ENST00000445902.2 |
VPS52
|
vacuolar protein sorting 52 homolog (S. cerevisiae) |
| chr9_+_124088860 | 0.28 |
ENST00000373806.1
|
GSN
|
gelsolin |
| chr3_+_62304648 | 0.28 |
ENST00000462069.1
ENST00000232519.5 ENST00000465142.1 |
C3orf14
|
chromosome 3 open reading frame 14 |
| chr20_+_55926625 | 0.28 |
ENST00000452119.1
|
RAE1
|
ribonucleic acid export 1 |
| chr19_-_55919087 | 0.28 |
ENST00000587845.1
ENST00000589978.1 ENST00000264552.9 |
UBE2S
|
ubiquitin-conjugating enzyme E2S |
| chr3_-_96337000 | 0.28 |
ENST00000600213.2
|
MTRNR2L12
|
MT-RNR2-like 12 (pseudogene) |
| chr2_-_106054952 | 0.28 |
ENST00000336660.5
ENST00000393352.3 ENST00000607522.1 |
FHL2
|
four and a half LIM domains 2 |
| chr17_+_80317121 | 0.28 |
ENST00000333437.4
|
TEX19
|
testis expressed 19 |
| chr3_+_142342228 | 0.28 |
ENST00000337777.3
|
PLS1
|
plastin 1 |
| chr17_+_38497640 | 0.28 |
ENST00000394086.3
|
RARA
|
retinoic acid receptor, alpha |
| chr7_-_30066233 | 0.28 |
ENST00000222803.5
|
FKBP14
|
FK506 binding protein 14, 22 kDa |
| chr16_-_2014804 | 0.27 |
ENST00000526522.1
ENST00000527302.1 ENST00000529806.1 ENST00000563194.1 ENST00000343262.4 |
RPS2
|
ribosomal protein S2 |
| chr3_+_57882061 | 0.27 |
ENST00000461354.1
ENST00000466255.1 |
SLMAP
|
sarcolemma associated protein |
| chr9_-_118417 | 0.27 |
ENST00000382500.2
|
FOXD4
|
forkhead box D4 |
| chr3_-_129035120 | 0.27 |
ENST00000333762.4
|
H1FX
|
H1 histone family, member X |
| chr6_+_159290917 | 0.26 |
ENST00000367072.1
|
C6orf99
|
chromosome 6 open reading frame 99 |
| chr6_+_10748019 | 0.26 |
ENST00000543878.1
ENST00000461342.1 ENST00000475942.1 ENST00000379530.3 ENST00000473276.1 ENST00000481240.1 ENST00000467317.1 |
SYCP2L
TMEM14B
|
synaptonemal complex protein 2-like transmembrane protein 14B |
| chr6_+_36562132 | 0.26 |
ENST00000373715.6
ENST00000339436.7 |
SRSF3
|
serine/arginine-rich splicing factor 3 |
| chr22_-_19466454 | 0.26 |
ENST00000494054.1
|
UFD1L
|
ubiquitin fusion degradation 1 like (yeast) |
| chr8_-_143696833 | 0.26 |
ENST00000356613.2
|
ARC
|
activity-regulated cytoskeleton-associated protein |
| chr15_+_49715293 | 0.26 |
ENST00000267843.4
ENST00000560270.1 |
FGF7
|
fibroblast growth factor 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TBP
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 5.4 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.5 | 1.5 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.4 | 3.9 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.4 | 1.2 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 0.4 | 3.1 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.3 | 1.0 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.3 | 1.4 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) |
| 0.3 | 0.8 | GO:0036023 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.2 | 0.6 | GO:0051037 | regulation of transcription involved in meiotic cell cycle(GO:0051037) |
| 0.2 | 1.4 | GO:0060356 | leucine import(GO:0060356) |
| 0.2 | 0.6 | GO:0007439 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.2 | 0.7 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.2 | 0.5 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) |
| 0.2 | 0.5 | GO:0035910 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) negative regulation of auditory receptor cell differentiation(GO:0045608) negative regulation of mechanoreceptor differentiation(GO:0045632) regulation of timing of neuron differentiation(GO:0060164) negative regulation of inner ear receptor cell differentiation(GO:2000981) |
| 0.2 | 1.9 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.1 | 0.4 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.1 | 0.7 | GO:0019401 | alditol biosynthetic process(GO:0019401) |
| 0.1 | 0.7 | GO:2000330 | positive regulation of T-helper 17 cell lineage commitment(GO:2000330) |
| 0.1 | 0.4 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.1 | 0.4 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.1 | 0.9 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.1 | 1.0 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.1 | 1.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.1 | 2.2 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.1 | 0.1 | GO:1903989 | positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
| 0.1 | 0.9 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 0.6 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.1 | 0.4 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.1 | 0.5 | GO:0002501 | MHC protein complex assembly(GO:0002396) peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.1 | 0.3 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.1 | 0.4 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.1 | 0.4 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 1.4 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.8 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.1 | 1.4 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.1 | 0.3 | GO:1900133 | regulation of renin secretion into blood stream(GO:1900133) |
| 0.1 | 0.3 | GO:2000224 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.1 | 0.2 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 0.3 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.1 | 0.4 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 0.1 | 0.3 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.1 | 0.6 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.1 | 0.3 | GO:1900154 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.1 | 0.3 | GO:2000969 | positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.1 | 0.2 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.1 | 0.4 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 0.2 | GO:1903515 | calcium ion transport from cytosol to endoplasmic reticulum(GO:1903515) |
| 0.1 | 0.5 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.1 | 0.3 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 0.3 | GO:0044856 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.1 | 0.3 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 0.2 | GO:0060437 | lung growth(GO:0060437) |
| 0.1 | 0.4 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 0.7 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.1 | 0.5 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.4 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.1 | 0.5 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.4 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 1.7 | GO:0050716 | positive regulation of interleukin-1 secretion(GO:0050716) |
| 0.0 | 0.2 | GO:0052042 | induction of programmed cell death(GO:0012502) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) |
| 0.0 | 0.1 | GO:0030474 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.0 | 0.6 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.5 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 5.4 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.1 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.0 | 0.1 | GO:0045083 | negative regulation of interleukin-12 biosynthetic process(GO:0045083) |
| 0.0 | 0.9 | GO:0030903 | notochord development(GO:0030903) |
| 0.0 | 0.4 | GO:1902669 | positive regulation of axon guidance(GO:1902669) |
| 0.0 | 0.3 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:0086092 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) |
| 0.0 | 0.2 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.1 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.4 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.0 | 0.6 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.0 | 0.2 | GO:0015853 | adenine transport(GO:0015853) |
| 0.0 | 0.4 | GO:0060347 | heart trabecula formation(GO:0060347) |
| 0.0 | 0.6 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.2 | GO:0046125 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.0 | 0.3 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.3 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.1 | GO:0090381 | negative regulation of mesodermal cell fate specification(GO:0042662) motor learning(GO:0061743) regulation of heart induction(GO:0090381) regulation of heart induction by canonical Wnt signaling pathway(GO:0100012) |
| 0.0 | 0.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.9 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.5 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.0 | GO:1904666 | regulation of ubiquitin protein ligase activity(GO:1904666) positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 0.1 | GO:0035900 | response to isolation stress(GO:0035900) |
| 0.0 | 0.4 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.3 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 | 0.1 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.1 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.5 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.0 | 0.5 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.4 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.0 | 0.2 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 1.1 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 2.2 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.2 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.0 | 0.6 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.2 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.1 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.5 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.2 | GO:1900120 | regulation of receptor binding(GO:1900120) |
| 0.0 | 0.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.4 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.4 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.0 | 0.4 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0050893 | sensory processing(GO:0050893) |
| 0.0 | 0.4 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.2 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.0 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.0 | GO:0018277 | protein deamination(GO:0018277) |
| 0.0 | 0.1 | GO:0030913 | paranodal junction assembly(GO:0030913) |
| 0.0 | 0.0 | GO:1903423 | synaptic vesicle recycling via endosome(GO:0036466) positive regulation of synaptic vesicle endocytosis(GO:1900244) positive regulation of synaptic vesicle recycling(GO:1903423) positive regulation of synaptic vesicle exocytosis(GO:2000302) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.8 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.3 | 14.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.2 | 0.5 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.2 | 0.6 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 1.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 0.7 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 2.4 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.1 | 0.5 | GO:0031906 | late endosome lumen(GO:0031906) |
| 0.1 | 0.3 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.1 | 1.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 4.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.2 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) |
| 0.1 | 3.5 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 1.5 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 0.3 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.1 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.0 | 1.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.5 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.7 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.4 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.5 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 1.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.5 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.2 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.1 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 0.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.6 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 2.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.1 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.4 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.2 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 1.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.4 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.2 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.3 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.2 | GO:0005882 | intermediate filament(GO:0005882) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.8 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.3 | 2.0 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.2 | 0.7 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.2 | 1.1 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.2 | 1.5 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.2 | 0.7 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.2 | 0.5 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.1 | 0.6 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.1 | 0.4 | GO:0080130 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 0.4 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.1 | 0.4 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.1 | 0.7 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.1 | 0.9 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.4 | GO:0031403 | lithium ion binding(GO:0031403) |
| 0.1 | 0.3 | GO:0047017 | geranylgeranyl reductase activity(GO:0045550) prostaglandin-F synthase activity(GO:0047017) |
| 0.1 | 0.3 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.1 | 0.6 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 0.5 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.1 | 1.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 3.1 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.5 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.2 | GO:0086039 | lutropin-choriogonadotropic hormone receptor binding(GO:0031775) calcium-transporting ATPase activity involved in regulation of cardiac muscle cell membrane potential(GO:0086039) |
| 0.1 | 0.3 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.2 | GO:0019828 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) aspartic-type endopeptidase inhibitor activity(GO:0019828) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 1.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 1.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.6 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0086040 | sodium:proton antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086040) |
| 0.0 | 1.2 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.1 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.0 | 0.1 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 0.3 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 0.2 | GO:0015207 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.0 | 0.3 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.4 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.3 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.4 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.1 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.0 | 0.8 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.2 | GO:0004797 | thymidine kinase activity(GO:0004797) |
| 0.0 | 0.6 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.3 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.3 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.6 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 3.8 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.1 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.0 | 0.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.6 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.1 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.0 | 0.7 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 1.1 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.3 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.3 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 1.2 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.3 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.3 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.9 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.1 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.0 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.6 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.2 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.1 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.4 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.5 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.2 | GO:0010857 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.0 | 0.4 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 7.8 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.3 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.0 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.2 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.0 | GO:0051400 | BH domain binding(GO:0051400) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 3.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.7 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 2.0 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.6 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.8 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 1.3 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.4 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.3 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 1.2 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.4 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.9 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.5 | PID TRAIL PATHWAY | TRAIL signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 28.3 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.4 | 7.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.2 | 1.5 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 2.0 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 1.7 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 1.9 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 2.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 2.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 0.9 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 1.4 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.4 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.5 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.3 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.3 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.4 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.8 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.2 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 0.4 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.5 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |