Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
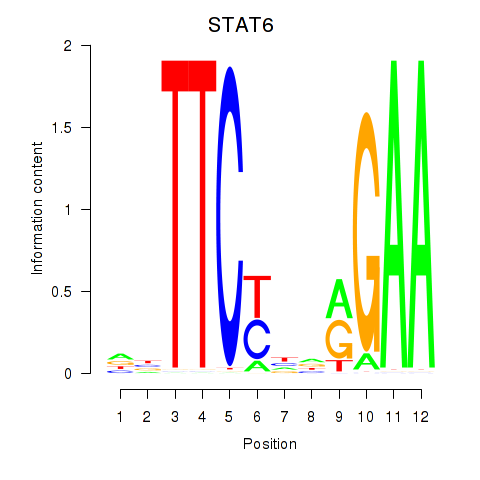
Results for STAT6
Z-value: 0.66
Transcription factors associated with STAT6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
STAT6
|
ENSG00000166888.6 | signal transducer and activator of transcription 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| STAT6 | hg19_v2_chr12_-_57505121_57505216 | -0.47 | 5.3e-01 | Click! |
Activity profile of STAT6 motif
Sorted Z-values of STAT6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_3469181 | 0.85 |
ENST00000366116.2
|
AL365202.1
|
Uncharacterized protein |
| chr12_-_122658746 | 0.41 |
ENST00000377035.1
|
IL31
|
interleukin 31 |
| chr6_+_138188351 | 0.32 |
ENST00000421450.1
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr18_+_66382428 | 0.31 |
ENST00000578970.1
ENST00000582371.1 ENST00000584775.1 |
CCDC102B
|
coiled-coil domain containing 102B |
| chr14_-_24732738 | 0.28 |
ENST00000558074.1
ENST00000560226.1 |
TGM1
|
transglutaminase 1 |
| chr5_+_667759 | 0.26 |
ENST00000594226.1
|
AC026740.1
|
Uncharacterized protein |
| chr3_+_72201910 | 0.26 |
ENST00000469178.1
ENST00000485404.1 |
LINC00870
|
long intergenic non-protein coding RNA 870 |
| chr8_+_97506033 | 0.22 |
ENST00000518385.1
|
SDC2
|
syndecan 2 |
| chr17_-_34207295 | 0.21 |
ENST00000463941.1
ENST00000293272.3 |
CCL5
|
chemokine (C-C motif) ligand 5 |
| chr6_+_30951487 | 0.20 |
ENST00000486149.2
ENST00000376296.3 |
MUC21
|
mucin 21, cell surface associated |
| chr8_+_39972170 | 0.20 |
ENST00000521257.1
|
RP11-359E19.2
|
RP11-359E19.2 |
| chr1_+_149754227 | 0.20 |
ENST00000444948.1
ENST00000369168.4 |
FCGR1A
|
Fc fragment of IgG, high affinity Ia, receptor (CD64) |
| chrX_-_139866723 | 0.19 |
ENST00000370532.2
|
CDR1
|
cerebellar degeneration-related protein 1, 34kDa |
| chr17_+_40912764 | 0.17 |
ENST00000589683.1
ENST00000588928.1 |
RAMP2
|
receptor (G protein-coupled) activity modifying protein 2 |
| chr5_+_112227311 | 0.17 |
ENST00000391338.1
|
ZRSR1
|
zinc finger (CCCH type), RNA-binding motif and serine/arginine rich 1 |
| chrX_+_135251835 | 0.17 |
ENST00000456445.1
|
FHL1
|
four and a half LIM domains 1 |
| chr17_+_7384721 | 0.17 |
ENST00000412468.2
|
SLC35G6
|
solute carrier family 35, member G6 |
| chr5_-_74162739 | 0.17 |
ENST00000513277.1
|
FAM169A
|
family with sequence similarity 169, member A |
| chr17_+_28268623 | 0.17 |
ENST00000394835.3
ENST00000320856.5 ENST00000394832.2 ENST00000378738.3 |
EFCAB5
|
EF-hand calcium binding domain 5 |
| chr12_-_111358372 | 0.16 |
ENST00000548438.1
ENST00000228841.8 |
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow |
| chr11_+_76745385 | 0.15 |
ENST00000533140.1
ENST00000354301.5 ENST00000528622.1 |
B3GNT6
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 6 (core 3 synthase) |
| chrX_+_135252050 | 0.15 |
ENST00000449474.1
ENST00000345434.3 |
FHL1
|
four and a half LIM domains 1 |
| chr3_-_98235962 | 0.14 |
ENST00000513873.1
|
CLDND1
|
claudin domain containing 1 |
| chrX_+_135251783 | 0.14 |
ENST00000394153.2
|
FHL1
|
four and a half LIM domains 1 |
| chr11_-_14379997 | 0.14 |
ENST00000526063.1
ENST00000532814.1 |
RRAS2
|
related RAS viral (r-ras) oncogene homolog 2 |
| chr17_+_32582293 | 0.14 |
ENST00000580907.1
ENST00000225831.4 |
CCL2
|
chemokine (C-C motif) ligand 2 |
| chr7_+_76091775 | 0.13 |
ENST00000442516.1
|
DTX2
|
deltex homolog 2 (Drosophila) |
| chr17_-_39681578 | 0.12 |
ENST00000593096.1
|
KRT19
|
keratin 19 |
| chr7_+_112120908 | 0.12 |
ENST00000439068.2
ENST00000312849.4 ENST00000429049.1 |
LSMEM1
|
leucine-rich single-pass membrane protein 1 |
| chr3_+_140981456 | 0.12 |
ENST00000504264.1
|
ACPL2
|
acid phosphatase-like 2 |
| chr6_-_41168920 | 0.12 |
ENST00000483722.1
|
TREML2
|
triggering receptor expressed on myeloid cells-like 2 |
| chr12_-_46663734 | 0.11 |
ENST00000550173.1
|
SLC38A1
|
solute carrier family 38, member 1 |
| chr3_+_10312604 | 0.11 |
ENST00000426850.1
|
TATDN2
|
TatD DNase domain containing 2 |
| chr12_+_7053228 | 0.11 |
ENST00000540506.2
|
C12orf57
|
chromosome 12 open reading frame 57 |
| chr16_+_30075595 | 0.10 |
ENST00000563060.2
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr5_-_146461027 | 0.10 |
ENST00000394410.2
ENST00000508267.1 ENST00000504198.1 |
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr16_+_30075783 | 0.10 |
ENST00000412304.2
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr13_+_50589390 | 0.09 |
ENST00000360473.4
ENST00000312942.1 |
KCNRG
|
potassium channel regulator |
| chr11_-_119252359 | 0.09 |
ENST00000455332.2
|
USP2
|
ubiquitin specific peptidase 2 |
| chr22_+_44464923 | 0.09 |
ENST00000404989.1
|
PARVB
|
parvin, beta |
| chr17_-_48277552 | 0.09 |
ENST00000507689.1
|
COL1A1
|
collagen, type I, alpha 1 |
| chr16_+_57728701 | 0.09 |
ENST00000569375.1
ENST00000360716.3 ENST00000569167.1 ENST00000394337.4 ENST00000563126.1 ENST00000336825.8 |
CCDC135
|
coiled-coil domain containing 135 |
| chr16_+_69985083 | 0.09 |
ENST00000288040.6
ENST00000449317.2 |
CLEC18A
|
C-type lectin domain family 18, member A |
| chr12_+_7053172 | 0.08 |
ENST00000229281.5
|
C12orf57
|
chromosome 12 open reading frame 57 |
| chr3_-_45957088 | 0.08 |
ENST00000539217.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr16_-_67694597 | 0.08 |
ENST00000393919.4
ENST00000219251.8 |
ACD
|
adrenocortical dysplasia homolog (mouse) |
| chr16_+_69984810 | 0.08 |
ENST00000393701.2
ENST00000568461.1 |
CLEC18A
|
C-type lectin domain family 18, member A |
| chr7_-_75401513 | 0.08 |
ENST00000005180.4
|
CCL26
|
chemokine (C-C motif) ligand 26 |
| chr4_-_120988229 | 0.08 |
ENST00000296509.6
|
MAD2L1
|
MAD2 mitotic arrest deficient-like 1 (yeast) |
| chr7_+_138818490 | 0.08 |
ENST00000430935.1
ENST00000495038.1 ENST00000474035.2 ENST00000478836.2 ENST00000464848.1 ENST00000343187.4 |
TTC26
|
tetratricopeptide repeat domain 26 |
| chr2_-_197226875 | 0.08 |
ENST00000409111.1
|
HECW2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr11_-_119252425 | 0.08 |
ENST00000260187.2
|
USP2
|
ubiquitin specific peptidase 2 |
| chr9_-_111619239 | 0.08 |
ENST00000374667.3
|
ACTL7B
|
actin-like 7B |
| chr11_+_36616044 | 0.07 |
ENST00000334307.5
ENST00000531554.1 ENST00000347206.4 ENST00000534635.1 ENST00000446510.2 ENST00000530697.1 ENST00000527108.1 |
C11orf74
|
chromosome 11 open reading frame 74 |
| chr6_-_24877490 | 0.07 |
ENST00000540914.1
ENST00000378023.4 |
FAM65B
|
family with sequence similarity 65, member B |
| chr18_+_33161698 | 0.07 |
ENST00000591924.1
|
GALNT1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 (GalNAc-T1) |
| chr8_-_82359662 | 0.07 |
ENST00000519260.1
ENST00000256103.2 |
PMP2
|
peripheral myelin protein 2 |
| chr8_+_11666649 | 0.07 |
ENST00000528643.1
ENST00000525777.1 |
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr2_-_183731882 | 0.07 |
ENST00000295113.4
|
FRZB
|
frizzled-related protein |
| chr22_-_42342733 | 0.07 |
ENST00000402420.1
|
CENPM
|
centromere protein M |
| chr2_-_31440377 | 0.07 |
ENST00000444918.2
ENST00000403897.3 |
CAPN14
|
calpain 14 |
| chr5_+_118691706 | 0.07 |
ENST00000415806.2
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr16_+_30075463 | 0.07 |
ENST00000562168.1
ENST00000569545.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr2_+_202316392 | 0.07 |
ENST00000194530.3
ENST00000392249.2 |
STRADB
|
STE20-related kinase adaptor beta |
| chr1_+_12123414 | 0.07 |
ENST00000263932.2
|
TNFRSF8
|
tumor necrosis factor receptor superfamily, member 8 |
| chr18_-_53068782 | 0.06 |
ENST00000569012.1
|
TCF4
|
transcription factor 4 |
| chr2_+_191792376 | 0.06 |
ENST00000409428.1
ENST00000409215.1 |
GLS
|
glutaminase |
| chr1_+_174417095 | 0.06 |
ENST00000367685.2
|
GPR52
|
G protein-coupled receptor 52 |
| chr2_+_103378472 | 0.06 |
ENST00000412401.2
|
TMEM182
|
transmembrane protein 182 |
| chr7_-_150777920 | 0.06 |
ENST00000353841.2
ENST00000297532.6 |
FASTK
|
Fas-activated serine/threonine kinase |
| chr10_+_115674530 | 0.06 |
ENST00000451472.1
|
AL162407.1
|
CDNA FLJ20147 fis, clone COL07954; HCG1781466; Uncharacterized protein |
| chr15_+_27111510 | 0.05 |
ENST00000335625.5
|
GABRA5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr20_-_23066953 | 0.05 |
ENST00000246006.4
|
CD93
|
CD93 molecule |
| chr2_-_203735484 | 0.05 |
ENST00000420558.1
ENST00000418208.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr4_-_46391931 | 0.05 |
ENST00000381620.4
|
GABRA2
|
gamma-aminobutyric acid (GABA) A receptor, alpha 2 |
| chr2_+_173600565 | 0.05 |
ENST00000397081.3
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr19_+_15218180 | 0.05 |
ENST00000342784.2
ENST00000597977.1 ENST00000600440.1 |
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans) |
| chr6_+_138188378 | 0.05 |
ENST00000420009.1
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr20_+_42574317 | 0.05 |
ENST00000358131.5
|
TOX2
|
TOX high mobility group box family member 2 |
| chr2_+_68961905 | 0.05 |
ENST00000295381.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chrX_+_102192200 | 0.05 |
ENST00000218249.5
|
RAB40AL
|
RAB40A, member RAS oncogene family-like |
| chr2_+_68961934 | 0.05 |
ENST00000409202.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr6_-_27860956 | 0.05 |
ENST00000359611.2
|
HIST1H2AM
|
histone cluster 1, H2am |
| chr2_+_173600514 | 0.05 |
ENST00000264111.6
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr11_-_60719213 | 0.05 |
ENST00000227880.3
|
SLC15A3
|
solute carrier family 15 (oligopeptide transporter), member 3 |
| chr19_+_13228917 | 0.05 |
ENST00000586171.1
|
NACC1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr4_+_30721968 | 0.05 |
ENST00000361762.2
|
PCDH7
|
protocadherin 7 |
| chr17_+_7461613 | 0.04 |
ENST00000438470.1
ENST00000436057.1 |
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr11_+_63655987 | 0.04 |
ENST00000509502.2
ENST00000512060.1 |
MARK2
|
MAP/microtubule affinity-regulating kinase 2 |
| chr3_+_41236325 | 0.04 |
ENST00000426215.1
ENST00000405570.1 |
CTNNB1
|
catenin (cadherin-associated protein), beta 1, 88kDa |
| chr17_+_68071389 | 0.04 |
ENST00000283936.1
ENST00000392671.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr2_-_204400113 | 0.04 |
ENST00000319170.5
|
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr15_+_96869165 | 0.04 |
ENST00000421109.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr2_-_204399976 | 0.04 |
ENST00000457812.1
|
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr21_+_17791838 | 0.04 |
ENST00000453910.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr4_+_57276661 | 0.04 |
ENST00000598320.1
|
AC068620.1
|
Uncharacterized protein |
| chr16_-_74455290 | 0.04 |
ENST00000339953.5
|
CLEC18B
|
C-type lectin domain family 18, member B |
| chr19_+_2785458 | 0.04 |
ENST00000307741.6
ENST00000585338.1 |
THOP1
|
thimet oligopeptidase 1 |
| chr1_+_26759295 | 0.04 |
ENST00000430232.1
|
DHDDS
|
dehydrodolichyl diphosphate synthase |
| chr10_-_35379241 | 0.04 |
ENST00000374748.1
ENST00000374749.3 |
CUL2
|
cullin 2 |
| chr3_-_45957534 | 0.04 |
ENST00000536047.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr16_-_29757272 | 0.04 |
ENST00000329410.3
|
C16orf54
|
chromosome 16 open reading frame 54 |
| chr5_+_102201687 | 0.04 |
ENST00000304400.7
|
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr9_-_100000957 | 0.04 |
ENST00000366109.2
ENST00000607322.1 |
RP11-498P14.5
|
RP11-498P14.5 |
| chr2_-_203736150 | 0.04 |
ENST00000457524.1
ENST00000421334.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr5_+_138609441 | 0.04 |
ENST00000509990.1
ENST00000506147.1 ENST00000512107.1 |
MATR3
|
matrin 3 |
| chr14_-_69446034 | 0.04 |
ENST00000193403.6
|
ACTN1
|
actinin, alpha 1 |
| chr1_+_109265033 | 0.04 |
ENST00000445274.1
|
FNDC7
|
fibronectin type III domain containing 7 |
| chrX_-_72434628 | 0.04 |
ENST00000536638.1
ENST00000373517.3 |
NAP1L2
|
nucleosome assembly protein 1-like 2 |
| chr12_+_7052974 | 0.03 |
ENST00000544681.1
ENST00000537087.1 |
C12orf57
|
chromosome 12 open reading frame 57 |
| chr8_-_37594944 | 0.03 |
ENST00000330539.1
|
RP11-863K10.7
|
Uncharacterized protein |
| chr2_+_68962014 | 0.03 |
ENST00000467265.1
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr3_+_52448539 | 0.03 |
ENST00000461861.1
|
PHF7
|
PHD finger protein 7 |
| chr1_-_8086343 | 0.03 |
ENST00000474874.1
ENST00000469499.1 ENST00000377482.5 |
ERRFI1
|
ERBB receptor feedback inhibitor 1 |
| chr1_+_66796401 | 0.03 |
ENST00000528771.1
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr3_-_49851313 | 0.03 |
ENST00000333486.3
|
UBA7
|
ubiquitin-like modifier activating enzyme 7 |
| chr19_+_38924316 | 0.03 |
ENST00000355481.4
ENST00000360985.3 ENST00000359596.3 |
RYR1
|
ryanodine receptor 1 (skeletal) |
| chr1_-_209824643 | 0.03 |
ENST00000391911.1
ENST00000415782.1 |
LAMB3
|
laminin, beta 3 |
| chr15_-_64386120 | 0.03 |
ENST00000300030.3
|
FAM96A
|
family with sequence similarity 96, member A |
| chr6_-_11232891 | 0.03 |
ENST00000379433.5
ENST00000379446.5 |
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr11_-_60720002 | 0.03 |
ENST00000538739.1
|
SLC15A3
|
solute carrier family 15 (oligopeptide transporter), member 3 |
| chr12_-_118490217 | 0.03 |
ENST00000542304.1
|
WSB2
|
WD repeat and SOCS box containing 2 |
| chr22_+_32439019 | 0.03 |
ENST00000266088.4
|
SLC5A1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr2_+_120124497 | 0.03 |
ENST00000355857.3
ENST00000535617.1 ENST00000535757.1 ENST00000409094.1 ENST00000311521.4 |
DBI
|
diazepam binding inhibitor (GABA receptor modulator, acyl-CoA binding protein) |
| chr11_-_128457446 | 0.03 |
ENST00000392668.4
|
ETS1
|
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
| chr11_+_112832090 | 0.03 |
ENST00000533760.1
|
NCAM1
|
neural cell adhesion molecule 1 |
| chr1_+_32608566 | 0.03 |
ENST00000545542.1
|
KPNA6
|
karyopherin alpha 6 (importin alpha 7) |
| chr2_+_234526272 | 0.03 |
ENST00000373450.4
|
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr18_-_21017817 | 0.03 |
ENST00000542162.1
ENST00000383233.3 ENST00000582336.1 ENST00000450466.2 ENST00000578520.1 ENST00000399707.1 |
TMEM241
|
transmembrane protein 241 |
| chr6_+_10556215 | 0.03 |
ENST00000316170.3
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr17_+_7462103 | 0.03 |
ENST00000396545.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr10_-_35379524 | 0.03 |
ENST00000374751.3
ENST00000374742.1 ENST00000602371.1 |
CUL2
|
cullin 2 |
| chr10_-_61122220 | 0.03 |
ENST00000422313.2
ENST00000435852.2 ENST00000442566.3 ENST00000373868.2 ENST00000277705.6 ENST00000373867.3 ENST00000419214.2 |
FAM13C
|
family with sequence similarity 13, member C |
| chr14_+_55034599 | 0.03 |
ENST00000392067.3
ENST00000357634.3 |
SAMD4A
|
sterile alpha motif domain containing 4A |
| chr2_+_219745020 | 0.03 |
ENST00000258411.3
|
WNT10A
|
wingless-type MMTV integration site family, member 10A |
| chr12_-_118490403 | 0.03 |
ENST00000535496.1
|
WSB2
|
WD repeat and SOCS box containing 2 |
| chr12_-_54779511 | 0.02 |
ENST00000551109.1
ENST00000546970.1 |
ZNF385A
|
zinc finger protein 385A |
| chr11_-_58980342 | 0.02 |
ENST00000361050.3
|
MPEG1
|
macrophage expressed 1 |
| chr15_+_101402041 | 0.02 |
ENST00000558475.1
ENST00000558641.1 ENST00000559673.1 |
RP11-66B24.1
|
RP11-66B24.1 |
| chr5_-_88120151 | 0.02 |
ENST00000506716.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr11_+_112832133 | 0.02 |
ENST00000524665.1
|
NCAM1
|
neural cell adhesion molecule 1 |
| chr6_+_31730773 | 0.02 |
ENST00000415669.2
ENST00000425424.1 |
SAPCD1
|
suppressor APC domain containing 1 |
| chr4_+_71019903 | 0.02 |
ENST00000344526.5
|
C4orf40
|
chromosome 4 open reading frame 40 |
| chr11_+_112832202 | 0.02 |
ENST00000534015.1
|
NCAM1
|
neural cell adhesion molecule 1 |
| chr1_-_190446759 | 0.02 |
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr6_+_26440700 | 0.02 |
ENST00000494393.1
ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3
|
butyrophilin, subfamily 3, member A3 |
| chr10_+_75541796 | 0.02 |
ENST00000372837.3
ENST00000372833.5 |
CHCHD1
|
coiled-coil-helix-coiled-coil-helix domain containing 1 |
| chr12_+_56435637 | 0.02 |
ENST00000356464.5
ENST00000552361.1 |
RPS26
|
ribosomal protein S26 |
| chr21_-_35340759 | 0.02 |
ENST00000607953.1
|
AP000569.9
|
AP000569.9 |
| chr11_+_65101225 | 0.02 |
ENST00000528416.1
ENST00000415073.2 ENST00000252268.4 |
DPF2
|
D4, zinc and double PHD fingers family 2 |
| chr3_+_51575596 | 0.02 |
ENST00000409535.2
|
RAD54L2
|
RAD54-like 2 (S. cerevisiae) |
| chr11_-_8964580 | 0.02 |
ENST00000325884.1
|
ASCL3
|
achaete-scute family bHLH transcription factor 3 |
| chr3_+_132379154 | 0.02 |
ENST00000468022.1
ENST00000473651.1 ENST00000494238.2 |
UBA5
|
ubiquitin-like modifier activating enzyme 5 |
| chr2_-_120124258 | 0.02 |
ENST00000409877.1
ENST00000409523.1 ENST00000409466.2 ENST00000414534.1 |
C2orf76
|
chromosome 2 open reading frame 76 |
| chr1_-_94586651 | 0.02 |
ENST00000535735.1
ENST00000370225.3 |
ABCA4
|
ATP-binding cassette, sub-family A (ABC1), member 4 |
| chr1_+_109256067 | 0.02 |
ENST00000271311.2
|
FNDC7
|
fibronectin type III domain containing 7 |
| chr4_-_10686475 | 0.02 |
ENST00000226951.6
|
CLNK
|
cytokine-dependent hematopoietic cell linker |
| chr12_+_9144626 | 0.02 |
ENST00000543895.1
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr11_-_10828892 | 0.02 |
ENST00000525681.1
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr3_+_121311966 | 0.02 |
ENST00000338040.4
|
FBXO40
|
F-box protein 40 |
| chrX_-_106960285 | 0.02 |
ENST00000503515.1
ENST00000372397.2 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr22_-_42342692 | 0.02 |
ENST00000404067.1
ENST00000402338.1 |
CENPM
|
centromere protein M |
| chr12_+_6493319 | 0.02 |
ENST00000536876.1
|
LTBR
|
lymphotoxin beta receptor (TNFR superfamily, member 3) |
| chr14_+_56127989 | 0.01 |
ENST00000555573.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr1_-_205649580 | 0.01 |
ENST00000367145.3
|
SLC45A3
|
solute carrier family 45, member 3 |
| chr15_-_64385981 | 0.01 |
ENST00000557835.1
ENST00000380290.3 ENST00000559950.1 |
FAM96A
|
family with sequence similarity 96, member A |
| chr2_+_234686976 | 0.01 |
ENST00000389758.3
|
MROH2A
|
maestro heat-like repeat family member 2A |
| chr9_-_135996537 | 0.01 |
ENST00000372050.3
ENST00000372047.3 |
RALGDS
|
ral guanine nucleotide dissociation stimulator |
| chr3_+_189507460 | 0.01 |
ENST00000434928.1
|
TP63
|
tumor protein p63 |
| chr1_+_82165350 | 0.01 |
ENST00000359929.3
|
LPHN2
|
latrophilin 2 |
| chr14_+_59100774 | 0.01 |
ENST00000556859.1
ENST00000421793.1 |
DACT1
|
dishevelled-binding antagonist of beta-catenin 1 |
| chr21_+_42539701 | 0.01 |
ENST00000330333.6
ENST00000328735.6 ENST00000347667.5 |
BACE2
|
beta-site APP-cleaving enzyme 2 |
| chr5_+_35856951 | 0.01 |
ENST00000303115.3
ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr1_-_68299130 | 0.01 |
ENST00000370982.3
|
GNG12
|
guanine nucleotide binding protein (G protein), gamma 12 |
| chr5_+_74011328 | 0.01 |
ENST00000513336.1
|
HEXB
|
hexosaminidase B (beta polypeptide) |
| chr3_+_153839149 | 0.01 |
ENST00000465093.1
ENST00000465817.1 |
ARHGEF26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr1_+_89246647 | 0.01 |
ENST00000544045.1
|
PKN2
|
protein kinase N2 |
| chr5_+_93954358 | 0.01 |
ENST00000504099.1
|
ANKRD32
|
ankyrin repeat domain 32 |
| chr12_-_6451235 | 0.01 |
ENST00000440083.2
ENST00000162749.2 |
TNFRSF1A
|
tumor necrosis factor receptor superfamily, member 1A |
| chr2_-_202316169 | 0.01 |
ENST00000430254.1
|
TRAK2
|
trafficking protein, kinesin binding 2 |
| chr4_+_144312659 | 0.01 |
ENST00000509992.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr14_-_24553834 | 0.01 |
ENST00000397002.2
|
NRL
|
neural retina leucine zipper |
| chr20_-_55100981 | 0.01 |
ENST00000243913.4
|
GCNT7
|
glucosaminyl (N-acetyl) transferase family member 7 |
| chr3_+_189507432 | 0.01 |
ENST00000354600.5
|
TP63
|
tumor protein p63 |
| chr14_-_37641618 | 0.01 |
ENST00000555449.1
|
SLC25A21
|
solute carrier family 25 (mitochondrial oxoadipate carrier), member 21 |
| chr16_+_67694849 | 0.01 |
ENST00000602551.1
ENST00000458121.2 ENST00000219255.3 |
PARD6A
|
par-6 family cell polarity regulator alpha |
| chr2_-_133427767 | 0.01 |
ENST00000397463.2
|
LYPD1
|
LY6/PLAUR domain containing 1 |
| chr7_+_129015671 | 0.01 |
ENST00000466993.1
|
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr3_+_158991025 | 0.01 |
ENST00000337808.6
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr7_-_150777949 | 0.01 |
ENST00000482571.1
|
FASTK
|
Fas-activated serine/threonine kinase |
| chr4_+_110769258 | 0.01 |
ENST00000594814.1
|
LRIT3
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3 |
| chr12_-_71031185 | 0.01 |
ENST00000548122.1
ENST00000551525.1 ENST00000550358.1 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr20_-_18477862 | 0.01 |
ENST00000337227.4
|
RBBP9
|
retinoblastoma binding protein 9 |
| chr8_-_117886955 | 0.01 |
ENST00000297338.2
|
RAD21
|
RAD21 homolog (S. pombe) |
| chr17_-_62308087 | 0.01 |
ENST00000583097.1
|
TEX2
|
testis expressed 2 |
| chr1_-_116383738 | 0.01 |
ENST00000320238.3
|
NHLH2
|
nescient helix loop helix 2 |
| chr20_+_58251716 | 0.01 |
ENST00000355648.4
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr15_+_34638066 | 0.01 |
ENST00000333756.4
|
NUTM1
|
NUT midline carcinoma, family member 1 |
| chr10_+_124907638 | 0.01 |
ENST00000339992.3
|
HMX2
|
H6 family homeobox 2 |
| chr12_-_71031220 | 0.00 |
ENST00000334414.6
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
Network of associatons between targets according to the STRING database.
First level regulatory network of STAT6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0034147 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) negative regulation of CD40 signaling pathway(GO:2000349) |
| 0.1 | 0.2 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.3 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.1 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.0 | 0.2 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.0 | 0.1 | GO:1903567 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.0 | 0.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.0 | 0.2 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.1 | GO:0060381 | regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) |
| 0.0 | 0.1 | GO:0032679 | TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) |
| 0.0 | 0.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.2 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.1 | GO:0006696 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.0 | GO:0061324 | glial cell fate determination(GO:0007403) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.0 | 0.0 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.0 | 0.1 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.0 | 0.2 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.0 | 0.1 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.0 | 0.2 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.1 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.2 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.1 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.0 | 0.1 | GO:0004310 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.1 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.1 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |