Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for STAT1_STAT3_BCL6
Z-value: 0.58
Transcription factors associated with STAT1_STAT3_BCL6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
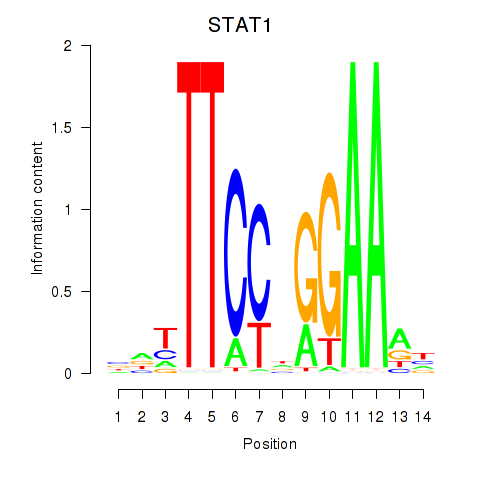
STAT1
|
ENSG00000115415.14 | signal transducer and activator of transcription 1 |
|
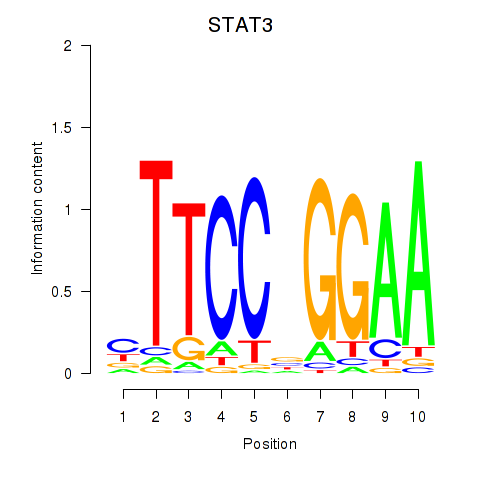
STAT3
|
ENSG00000168610.10 | signal transducer and activator of transcription 3 |
|
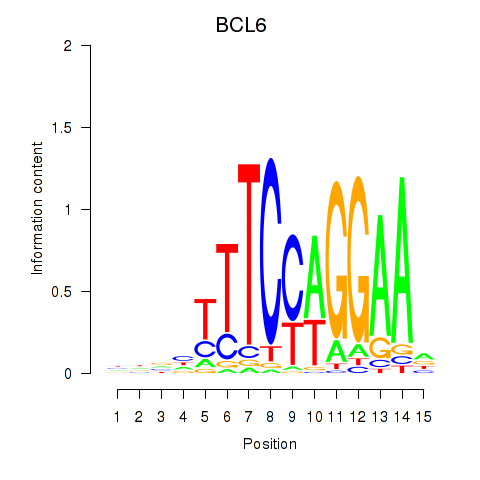
BCL6
|
ENSG00000113916.13 | BCL6 transcription repressor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BCL6 | hg19_v2_chr3_-_187455680_187455732 | 0.96 | 3.6e-02 | Click! |
| STAT3 | hg19_v2_chr17_-_40540586_40540600 | 0.60 | 4.0e-01 | Click! |
| STAT1 | hg19_v2_chr2_-_191885686_191885750 | 0.52 | 4.8e-01 | Click! |
Activity profile of STAT1_STAT3_BCL6 motif
Sorted Z-values of STAT1_STAT3_BCL6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_14396121 | 0.90 |
ENST00000570945.1
|
RP11-65J21.3
|
RP11-65J21.3 |
| chr3_-_187455680 | 0.71 |
ENST00000438077.1
|
BCL6
|
B-cell CLL/lymphoma 6 |
| chr22_-_27620603 | 0.48 |
ENST00000418271.1
ENST00000444114.1 |
RP5-1172A22.1
|
RP5-1172A22.1 |
| chr15_+_76030311 | 0.38 |
ENST00000543887.1
|
AC019294.1
|
AC019294.1 |
| chr14_-_80678512 | 0.31 |
ENST00000553968.1
|
DIO2
|
deiodinase, iodothyronine, type II |
| chr3_+_122399697 | 0.29 |
ENST00000494811.1
|
PARP14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr3_+_112930946 | 0.26 |
ENST00000462425.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr3_-_93692681 | 0.25 |
ENST00000348974.4
|
PROS1
|
protein S (alpha) |
| chr19_+_18530184 | 0.24 |
ENST00000601357.2
|
SSBP4
|
single stranded DNA binding protein 4 |
| chr19_-_40324767 | 0.24 |
ENST00000601972.1
ENST00000430012.2 ENST00000323039.5 ENST00000348817.3 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr17_-_42994283 | 0.23 |
ENST00000593179.1
|
GFAP
|
glial fibrillary acidic protein |
| chr14_-_81408063 | 0.23 |
ENST00000557411.1
|
CEP128
|
centrosomal protein 128kDa |
| chr17_+_76356516 | 0.23 |
ENST00000592569.1
|
RP11-806H10.4
|
RP11-806H10.4 |
| chr5_-_38845812 | 0.20 |
ENST00000513480.1
ENST00000512519.1 |
CTD-2127H9.1
|
CTD-2127H9.1 |
| chr19_-_40324255 | 0.19 |
ENST00000593685.1
ENST00000600611.1 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr14_+_94577074 | 0.18 |
ENST00000444961.1
ENST00000448882.1 ENST00000557098.1 ENST00000554800.1 ENST00000556544.1 ENST00000298902.5 ENST00000555819.1 ENST00000557634.1 ENST00000555744.1 |
IFI27
|
interferon, alpha-inducible protein 27 |
| chr3_-_69249863 | 0.18 |
ENST00000478263.1
ENST00000462512.1 |
FRMD4B
|
FERM domain containing 4B |
| chr19_-_49339915 | 0.18 |
ENST00000263278.4
|
HSD17B14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr1_-_168464875 | 0.17 |
ENST00000422253.1
|
RP5-968D22.3
|
RP5-968D22.3 |
| chr14_+_76452090 | 0.16 |
ENST00000314067.6
ENST00000238628.6 ENST00000556742.1 |
IFT43
|
intraflagellar transport 43 homolog (Chlamydomonas) |
| chr17_-_79917645 | 0.16 |
ENST00000477214.1
|
NOTUM
|
notum pectinacetylesterase homolog (Drosophila) |
| chr2_-_190044480 | 0.16 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr19_+_10196981 | 0.16 |
ENST00000591813.1
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr19_+_10381769 | 0.16 |
ENST00000423829.2
ENST00000588645.1 |
ICAM1
|
intercellular adhesion molecule 1 |
| chr19_-_58864848 | 0.16 |
ENST00000263100.3
|
A1BG
|
alpha-1-B glycoprotein |
| chr19_-_4831701 | 0.16 |
ENST00000248244.5
|
TICAM1
|
toll-like receptor adaptor molecule 1 |
| chr2_+_97203082 | 0.15 |
ENST00000454558.2
|
ARID5A
|
AT rich interactive domain 5A (MRF1-like) |
| chr19_+_57106624 | 0.15 |
ENST00000599599.1
|
ZNF71
|
zinc finger protein 71 |
| chr12_+_56325231 | 0.15 |
ENST00000549368.1
|
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr16_+_50280020 | 0.15 |
ENST00000564965.1
|
ADCY7
|
adenylate cyclase 7 |
| chr4_-_140477353 | 0.15 |
ENST00000406354.1
ENST00000506866.2 |
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr12_-_49245916 | 0.15 |
ENST00000552512.1
ENST00000551468.1 |
DDX23
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 23 |
| chr11_+_5710919 | 0.14 |
ENST00000379965.3
ENST00000425490.1 |
TRIM22
|
tripartite motif containing 22 |
| chr17_+_36858694 | 0.14 |
ENST00000563897.1
|
CTB-58E17.1
|
CTB-58E17.1 |
| chr1_-_153521714 | 0.14 |
ENST00000368713.3
|
S100A3
|
S100 calcium binding protein A3 |
| chr1_-_153521597 | 0.14 |
ENST00000368712.1
|
S100A3
|
S100 calcium binding protein A3 |
| chr1_-_230850043 | 0.14 |
ENST00000366667.4
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8) |
| chr17_+_6347761 | 0.14 |
ENST00000250056.8
ENST00000571373.1 ENST00000570337.2 ENST00000572595.2 ENST00000576056.1 |
FAM64A
|
family with sequence similarity 64, member A |
| chr2_+_87754989 | 0.13 |
ENST00000409898.2
ENST00000419680.2 ENST00000414584.1 ENST00000455131.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr1_-_41328018 | 0.13 |
ENST00000372638.2
|
CITED4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 |
| chr12_+_7022909 | 0.13 |
ENST00000537688.1
|
ENO2
|
enolase 2 (gamma, neuronal) |
| chr17_+_75315654 | 0.13 |
ENST00000590595.1
|
SEPT9
|
septin 9 |
| chr19_-_49339732 | 0.13 |
ENST00000599157.1
|
HSD17B14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr19_+_18530146 | 0.13 |
ENST00000348495.6
ENST00000270061.7 |
SSBP4
|
single stranded DNA binding protein 4 |
| chr11_+_76493294 | 0.13 |
ENST00000533752.1
|
TSKU
|
tsukushi, small leucine rich proteoglycan |
| chr1_+_221051699 | 0.12 |
ENST00000366903.6
|
HLX
|
H2.0-like homeobox |
| chr17_+_75276643 | 0.12 |
ENST00000589070.1
|
SEPT9
|
septin 9 |
| chr6_-_33548006 | 0.12 |
ENST00000374467.3
|
BAK1
|
BCL2-antagonist/killer 1 |
| chr7_-_26578407 | 0.12 |
ENST00000242109.3
|
KIAA0087
|
KIAA0087 |
| chr3_+_187871060 | 0.12 |
ENST00000448637.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr14_+_24779376 | 0.12 |
ENST00000530080.1
|
LTB4R2
|
leukotriene B4 receptor 2 |
| chr6_+_32821924 | 0.11 |
ENST00000374859.2
ENST00000453265.2 |
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr22_+_20748456 | 0.11 |
ENST00000420626.1
ENST00000356671.5 |
ZNF74
|
zinc finger protein 74 |
| chr6_+_108977520 | 0.11 |
ENST00000540898.1
|
FOXO3
|
forkhead box O3 |
| chrX_-_48931648 | 0.11 |
ENST00000376386.3
ENST00000376390.4 |
PRAF2
|
PRA1 domain family, member 2 |
| chr19_+_44617511 | 0.11 |
ENST00000262894.6
ENST00000588926.1 ENST00000592780.1 |
ZNF225
|
zinc finger protein 225 |
| chr12_-_57030096 | 0.11 |
ENST00000549506.1
|
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A |
| chr4_-_155511887 | 0.11 |
ENST00000302053.3
ENST00000403106.3 |
FGA
|
fibrinogen alpha chain |
| chr11_-_62609281 | 0.11 |
ENST00000525239.1
ENST00000538098.2 |
WDR74
|
WD repeat domain 74 |
| chr12_-_7261772 | 0.11 |
ENST00000545280.1
ENST00000543933.1 ENST00000545337.1 ENST00000544702.1 ENST00000266542.4 |
C1RL
|
complement component 1, r subcomponent-like |
| chr1_-_226065330 | 0.11 |
ENST00000436966.1
|
TMEM63A
|
transmembrane protein 63A |
| chr10_-_90751038 | 0.10 |
ENST00000458159.1
ENST00000415557.1 ENST00000458208.1 |
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr16_-_3030407 | 0.10 |
ENST00000431515.2
ENST00000574385.1 ENST00000576268.1 ENST00000574730.1 ENST00000575632.1 ENST00000573944.1 ENST00000262300.8 |
PKMYT1
|
protein kinase, membrane associated tyrosine/threonine 1 |
| chr15_-_88799661 | 0.10 |
ENST00000360948.2
ENST00000357724.2 ENST00000355254.2 ENST00000317501.3 |
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr12_-_7245018 | 0.10 |
ENST00000543835.1
ENST00000535233.2 |
C1R
|
complement component 1, r subcomponent |
| chr12_+_53443680 | 0.10 |
ENST00000314250.6
ENST00000451358.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr12_+_56415100 | 0.10 |
ENST00000547791.1
|
IKZF4
|
IKAROS family zinc finger 4 (Eos) |
| chr6_-_33547975 | 0.10 |
ENST00000442998.2
ENST00000360661.5 |
BAK1
|
BCL2-antagonist/killer 1 |
| chr13_-_34250861 | 0.10 |
ENST00000445227.1
ENST00000454681.2 |
RP11-141M1.3
|
RP11-141M1.3 |
| chr17_+_6347729 | 0.10 |
ENST00000572447.1
|
FAM64A
|
family with sequence similarity 64, member A |
| chr1_+_213123862 | 0.10 |
ENST00000366966.2
ENST00000366964.3 |
VASH2
|
vasohibin 2 |
| chr1_+_91966656 | 0.10 |
ENST00000428239.1
ENST00000426137.1 |
CDC7
|
cell division cycle 7 |
| chr1_+_10534944 | 0.10 |
ENST00000356607.4
ENST00000538836.1 ENST00000491661.2 |
PEX14
|
peroxisomal biogenesis factor 14 |
| chr12_+_122064398 | 0.10 |
ENST00000330079.7
|
ORAI1
|
ORAI calcium release-activated calcium modulator 1 |
| chr9_+_97766409 | 0.10 |
ENST00000425634.2
|
C9orf3
|
chromosome 9 open reading frame 3 |
| chr1_+_207262170 | 0.09 |
ENST00000367078.3
|
C4BPB
|
complement component 4 binding protein, beta |
| chr19_-_54974894 | 0.09 |
ENST00000333834.4
|
LENG9
|
leukocyte receptor cluster (LRC) member 9 |
| chr7_+_106415457 | 0.09 |
ENST00000490162.2
ENST00000470135.1 |
RP5-884M6.1
|
RP5-884M6.1 |
| chr19_-_41220540 | 0.09 |
ENST00000594490.1
|
ADCK4
|
aarF domain containing kinase 4 |
| chr14_+_100240019 | 0.09 |
ENST00000556199.1
|
EML1
|
echinoderm microtubule associated protein like 1 |
| chr8_-_97247759 | 0.09 |
ENST00000518406.1
ENST00000523920.1 ENST00000287022.5 |
UQCRB
|
ubiquinol-cytochrome c reductase binding protein |
| chr12_+_56324933 | 0.09 |
ENST00000549629.1
ENST00000555218.1 |
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr5_-_111092873 | 0.09 |
ENST00000509025.1
ENST00000515855.1 |
NREP
|
neuronal regeneration related protein |
| chr5_-_119669160 | 0.09 |
ENST00000514240.1
|
CTC-552D5.1
|
CTC-552D5.1 |
| chr12_-_11175219 | 0.09 |
ENST00000390673.2
|
TAS2R19
|
taste receptor, type 2, member 19 |
| chr19_+_15783879 | 0.09 |
ENST00000551607.1
|
CYP4F12
|
cytochrome P450, family 4, subfamily F, polypeptide 12 |
| chr22_+_24990746 | 0.09 |
ENST00000456869.1
ENST00000411974.1 |
GGT1
|
gamma-glutamyltransferase 1 |
| chr1_-_156571254 | 0.09 |
ENST00000438976.2
ENST00000334588.7 ENST00000368232.4 ENST00000415314.2 |
GPATCH4
|
G patch domain containing 4 |
| chr11_-_111649015 | 0.09 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr1_-_154946792 | 0.09 |
ENST00000412170.1
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr12_-_120687948 | 0.09 |
ENST00000458477.2
|
PXN
|
paxillin |
| chr12_-_42878101 | 0.09 |
ENST00000552108.1
|
PRICKLE1
|
prickle homolog 1 (Drosophila) |
| chr12_+_14561422 | 0.09 |
ENST00000541056.1
|
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr1_+_207262578 | 0.08 |
ENST00000243611.5
ENST00000367076.3 |
C4BPB
|
complement component 4 binding protein, beta |
| chr16_-_8955570 | 0.08 |
ENST00000567554.1
|
CARHSP1
|
calcium regulated heat stable protein 1, 24kDa |
| chr3_-_145940214 | 0.08 |
ENST00000481701.1
|
PLSCR4
|
phospholipid scramblase 4 |
| chr5_-_140013241 | 0.08 |
ENST00000519715.1
|
CD14
|
CD14 molecule |
| chr16_+_48657361 | 0.08 |
ENST00000565072.1
|
RP11-42I10.1
|
RP11-42I10.1 |
| chr5_+_40841410 | 0.08 |
ENST00000381677.3
|
CARD6
|
caspase recruitment domain family, member 6 |
| chr1_+_202431859 | 0.08 |
ENST00000391959.3
ENST00000367270.4 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr11_+_844067 | 0.08 |
ENST00000397406.1
ENST00000409543.2 ENST00000525201.1 |
TSPAN4
|
tetraspanin 4 |
| chr20_-_18774614 | 0.08 |
ENST00000412553.1
|
LINC00652
|
long intergenic non-protein coding RNA 652 |
| chr18_+_72166564 | 0.08 |
ENST00000583216.1
ENST00000581912.1 ENST00000582589.1 |
CNDP2
|
CNDP dipeptidase 2 (metallopeptidase M20 family) |
| chr3_-_120400960 | 0.08 |
ENST00000476082.2
|
HGD
|
homogentisate 1,2-dioxygenase |
| chr16_+_67563250 | 0.08 |
ENST00000566907.1
|
FAM65A
|
family with sequence similarity 65, member A |
| chr2_-_230787879 | 0.08 |
ENST00000435716.1
|
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr11_-_115127611 | 0.08 |
ENST00000545094.1
|
CADM1
|
cell adhesion molecule 1 |
| chr17_-_2304365 | 0.08 |
ENST00000575394.1
ENST00000174618.4 |
MNT
|
MAX network transcriptional repressor |
| chr19_+_44598492 | 0.08 |
ENST00000589680.1
|
ZNF224
|
zinc finger protein 224 |
| chr14_-_107283278 | 0.08 |
ENST00000390639.2
|
IGHV7-81
|
immunoglobulin heavy variable 7-81 (non-functional) |
| chr19_+_10197463 | 0.07 |
ENST00000590378.1
ENST00000397881.3 |
C19orf66
|
chromosome 19 open reading frame 66 |
| chr11_+_118754475 | 0.07 |
ENST00000292174.4
|
CXCR5
|
chemokine (C-X-C motif) receptor 5 |
| chr19_+_50145328 | 0.07 |
ENST00000360565.3
|
SCAF1
|
SR-related CTD-associated factor 1 |
| chr20_-_32580924 | 0.07 |
ENST00000432859.1
|
RP5-1125A11.1
|
RP5-1125A11.1 |
| chr6_-_127840048 | 0.07 |
ENST00000467753.1
|
SOGA3
|
SOGA family member 3 |
| chr19_+_17530838 | 0.07 |
ENST00000528659.1
ENST00000392702.2 ENST00000529939.1 |
MVB12A
|
multivesicular body subunit 12A |
| chr7_+_100728720 | 0.07 |
ENST00000306085.6
ENST00000412507.1 |
TRIM56
|
tripartite motif containing 56 |
| chr6_-_110011718 | 0.07 |
ENST00000532976.1
|
AK9
|
adenylate kinase 9 |
| chr2_-_183291741 | 0.07 |
ENST00000351439.5
ENST00000409365.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr5_-_146781153 | 0.07 |
ENST00000520473.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr1_-_151138323 | 0.07 |
ENST00000368908.5
|
LYSMD1
|
LysM, putative peptidoglycan-binding, domain containing 1 |
| chr22_-_39151463 | 0.07 |
ENST00000405510.1
ENST00000433561.1 |
SUN2
|
Sad1 and UNC84 domain containing 2 |
| chr16_-_57514277 | 0.07 |
ENST00000562008.1
ENST00000567214.1 |
DOK4
|
docking protein 4 |
| chr3_+_111393501 | 0.07 |
ENST00000393934.3
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr4_-_40632881 | 0.06 |
ENST00000511598.1
|
RBM47
|
RNA binding motif protein 47 |
| chr5_+_150639360 | 0.06 |
ENST00000523004.1
|
GM2A
|
GM2 ganglioside activator |
| chr20_+_42984330 | 0.06 |
ENST00000316673.4
ENST00000609795.1 ENST00000457232.1 ENST00000609262.1 |
HNF4A
|
hepatocyte nuclear factor 4, alpha |
| chr11_+_844406 | 0.06 |
ENST00000397404.1
|
TSPAN4
|
tetraspanin 4 |
| chr16_+_48657337 | 0.06 |
ENST00000568150.1
ENST00000564212.1 |
RP11-42I10.1
|
RP11-42I10.1 |
| chr19_+_50529329 | 0.06 |
ENST00000599155.1
|
ZNF473
|
zinc finger protein 473 |
| chr21_-_34915123 | 0.06 |
ENST00000438059.1
|
GART
|
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
| chr19_-_41220957 | 0.06 |
ENST00000596357.1
ENST00000243583.6 ENST00000600080.1 ENST00000595254.1 ENST00000601967.1 |
ADCK4
|
aarF domain containing kinase 4 |
| chr19_-_45927622 | 0.06 |
ENST00000300853.3
ENST00000589165.1 |
ERCC1
|
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) |
| chr1_+_16083098 | 0.06 |
ENST00000496928.2
ENST00000508310.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr2_+_17935383 | 0.06 |
ENST00000524465.1
ENST00000381254.2 ENST00000532257.1 |
GEN1
|
GEN1 Holliday junction 5' flap endonuclease |
| chr17_+_2699697 | 0.06 |
ENST00000254695.8
ENST00000366401.4 ENST00000542807.1 |
RAP1GAP2
|
RAP1 GTPase activating protein 2 |
| chr5_+_140579162 | 0.06 |
ENST00000536699.1
ENST00000354757.3 |
PCDHB11
|
protocadherin beta 11 |
| chr5_-_176738883 | 0.06 |
ENST00000513169.1
ENST00000423571.2 ENST00000502529.1 ENST00000427908.2 |
MXD3
|
MAX dimerization protein 3 |
| chr4_+_155484103 | 0.06 |
ENST00000302068.4
|
FGB
|
fibrinogen beta chain |
| chr3_-_139108463 | 0.06 |
ENST00000512242.1
|
COPB2
|
coatomer protein complex, subunit beta 2 (beta prime) |
| chr8_+_145726472 | 0.06 |
ENST00000528430.1
|
PPP1R16A
|
protein phosphatase 1, regulatory subunit 16A |
| chr12_-_58165870 | 0.06 |
ENST00000257848.7
|
METTL1
|
methyltransferase like 1 |
| chr1_+_207262627 | 0.06 |
ENST00000391923.1
|
C4BPB
|
complement component 4 binding protein, beta |
| chr17_+_19030782 | 0.06 |
ENST00000344415.4
ENST00000577213.1 |
GRAPL
|
GRB2-related adaptor protein-like |
| chr6_-_43484621 | 0.06 |
ENST00000506469.1
ENST00000503972.1 |
YIPF3
|
Yip1 domain family, member 3 |
| chr19_+_16186903 | 0.06 |
ENST00000588507.1
|
TPM4
|
tropomyosin 4 |
| chr4_-_48116540 | 0.06 |
ENST00000506073.1
|
TXK
|
TXK tyrosine kinase |
| chr17_-_6524159 | 0.06 |
ENST00000589033.1
|
KIAA0753
|
KIAA0753 |
| chr7_-_74267836 | 0.06 |
ENST00000361071.5
ENST00000453619.2 ENST00000417115.2 ENST00000405086.2 |
GTF2IRD2
|
GTF2I repeat domain containing 2 |
| chr22_+_20748405 | 0.06 |
ENST00000400451.2
ENST00000403682.3 ENST00000357502.5 |
ZNF74
|
zinc finger protein 74 |
| chr11_-_62323702 | 0.06 |
ENST00000530285.1
|
AHNAK
|
AHNAK nucleoprotein |
| chr11_-_111649074 | 0.06 |
ENST00000534218.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr3_-_137851220 | 0.06 |
ENST00000236709.3
|
A4GNT
|
alpha-1,4-N-acetylglucosaminyltransferase |
| chr19_-_10613361 | 0.06 |
ENST00000591039.1
ENST00000591419.1 |
KEAP1
|
kelch-like ECH-associated protein 1 |
| chr2_+_175352114 | 0.06 |
ENST00000444196.1
ENST00000417038.1 ENST00000606406.1 |
AC010894.3
|
AC010894.3 |
| chr12_+_53443963 | 0.06 |
ENST00000546602.1
ENST00000552570.1 ENST00000549700.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr1_+_42619070 | 0.06 |
ENST00000372581.1
|
GUCA2B
|
guanylate cyclase activator 2B (uroguanylin) |
| chr14_+_32030582 | 0.05 |
ENST00000550649.1
ENST00000281081.7 |
NUBPL
|
nucleotide binding protein-like |
| chr6_-_127840336 | 0.05 |
ENST00000525778.1
|
SOGA3
|
SOGA family member 3 |
| chr3_-_52864680 | 0.05 |
ENST00000406595.1
ENST00000485816.1 ENST00000434759.3 ENST00000346281.5 ENST00000266041.4 |
ITIH4
|
inter-alpha-trypsin inhibitor heavy chain family, member 4 |
| chr1_+_87794150 | 0.05 |
ENST00000370544.5
|
LMO4
|
LIM domain only 4 |
| chr19_-_44809121 | 0.05 |
ENST00000591609.1
ENST00000589799.1 ENST00000291182.4 ENST00000589248.1 |
ZNF235
|
zinc finger protein 235 |
| chr16_-_67260691 | 0.05 |
ENST00000447579.1
ENST00000393992.1 ENST00000424285.1 |
LRRC29
|
leucine rich repeat containing 29 |
| chr11_+_110001723 | 0.05 |
ENST00000528673.1
|
ZC3H12C
|
zinc finger CCCH-type containing 12C |
| chr9_+_97766469 | 0.05 |
ENST00000433691.2
|
C9orf3
|
chromosome 9 open reading frame 3 |
| chr6_-_127840021 | 0.05 |
ENST00000465909.2
|
SOGA3
|
SOGA family member 3 |
| chr6_+_35995488 | 0.05 |
ENST00000229795.3
|
MAPK14
|
mitogen-activated protein kinase 14 |
| chr19_+_15218180 | 0.05 |
ENST00000342784.2
ENST00000597977.1 ENST00000600440.1 |
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans) |
| chr7_-_150777874 | 0.05 |
ENST00000540185.1
|
FASTK
|
Fas-activated serine/threonine kinase |
| chr19_-_12845550 | 0.05 |
ENST00000242784.4
|
C19orf43
|
chromosome 19 open reading frame 43 |
| chr6_+_292253 | 0.05 |
ENST00000603453.1
ENST00000605315.1 ENST00000603881.1 |
DUSP22
|
dual specificity phosphatase 22 |
| chr6_-_52859046 | 0.05 |
ENST00000457564.1
ENST00000541324.1 ENST00000370960.1 |
GSTA4
|
glutathione S-transferase alpha 4 |
| chr7_+_2598628 | 0.05 |
ENST00000402050.2
ENST00000404984.1 ENST00000415271.2 ENST00000438376.2 |
IQCE
|
IQ motif containing E |
| chr19_+_13229126 | 0.05 |
ENST00000292431.4
|
NACC1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr17_-_1619568 | 0.05 |
ENST00000571595.1
|
MIR22HG
|
MIR22 host gene (non-protein coding) |
| chr8_-_144886321 | 0.05 |
ENST00000526832.1
|
SCRIB
|
scribbled planar cell polarity protein |
| chr19_-_47231216 | 0.05 |
ENST00000594287.2
|
STRN4
|
striatin, calmodulin binding protein 4 |
| chr1_+_1370903 | 0.05 |
ENST00000338660.5
ENST00000404702.3 ENST00000476993.1 ENST00000471398.1 |
VWA1
|
von Willebrand factor A domain containing 1 |
| chr7_-_100881109 | 0.05 |
ENST00000308344.5
|
CLDN15
|
claudin 15 |
| chr2_-_167232484 | 0.05 |
ENST00000375387.4
ENST00000303354.6 ENST00000409672.1 |
SCN9A
|
sodium channel, voltage-gated, type IX, alpha subunit |
| chr2_-_106013207 | 0.05 |
ENST00000447958.1
|
FHL2
|
four and a half LIM domains 2 |
| chr5_+_61708488 | 0.05 |
ENST00000505902.1
|
IPO11
|
importin 11 |
| chr19_+_49128209 | 0.05 |
ENST00000599748.1
ENST00000443164.1 ENST00000599029.1 |
SPHK2
|
sphingosine kinase 2 |
| chr3_+_185046676 | 0.05 |
ENST00000428617.1
ENST00000443863.1 |
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr19_-_7167989 | 0.05 |
ENST00000600492.1
|
INSR
|
insulin receptor |
| chr6_-_34524049 | 0.05 |
ENST00000374037.3
|
SPDEF
|
SAM pointed domain containing ETS transcription factor |
| chr3_+_160117062 | 0.05 |
ENST00000497311.1
|
SMC4
|
structural maintenance of chromosomes 4 |
| chr7_-_100881041 | 0.05 |
ENST00000412417.1
ENST00000414035.1 |
CLDN15
|
claudin 15 |
| chr16_-_29875057 | 0.05 |
ENST00000219789.6
|
CDIPT
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase |
| chr19_-_55672037 | 0.05 |
ENST00000588076.1
|
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr1_-_161207875 | 0.05 |
ENST00000512372.1
ENST00000437437.2 ENST00000442691.2 ENST00000412844.2 ENST00000428574.2 ENST00000505005.1 ENST00000508740.1 ENST00000508387.1 ENST00000504010.1 ENST00000511676.1 ENST00000502985.1 ENST00000367981.3 ENST00000515621.1 ENST00000511944.1 ENST00000511748.1 ENST00000367984.4 ENST00000367985.3 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr7_-_130353553 | 0.05 |
ENST00000330992.7
ENST00000445977.2 |
COPG2
|
coatomer protein complex, subunit gamma 2 |
| chr19_+_17416457 | 0.05 |
ENST00000252602.1
|
MRPL34
|
mitochondrial ribosomal protein L34 |
| chr1_+_40997233 | 0.05 |
ENST00000372699.3
ENST00000372697.3 ENST00000372696.3 |
ZNF684
|
zinc finger protein 684 |
| chr3_+_107364769 | 0.05 |
ENST00000449271.1
ENST00000425868.1 ENST00000449213.1 |
BBX
|
bobby sox homolog (Drosophila) |
| chr19_+_56905024 | 0.04 |
ENST00000591172.1
ENST00000589888.1 ENST00000587979.1 ENST00000585659.1 ENST00000593109.1 |
ZNF582-AS1
|
ZNF582 antisense RNA 1 (head to head) |
| chr19_+_42724423 | 0.04 |
ENST00000301215.3
ENST00000597945.1 |
ZNF526
|
zinc finger protein 526 |
| chr12_-_10007448 | 0.04 |
ENST00000538152.1
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr8_+_38586068 | 0.04 |
ENST00000443286.2
ENST00000520340.1 ENST00000518415.1 |
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr19_-_49250054 | 0.04 |
ENST00000602105.1
ENST00000332955.2 |
IZUMO1
|
izumo sperm-egg fusion 1 |
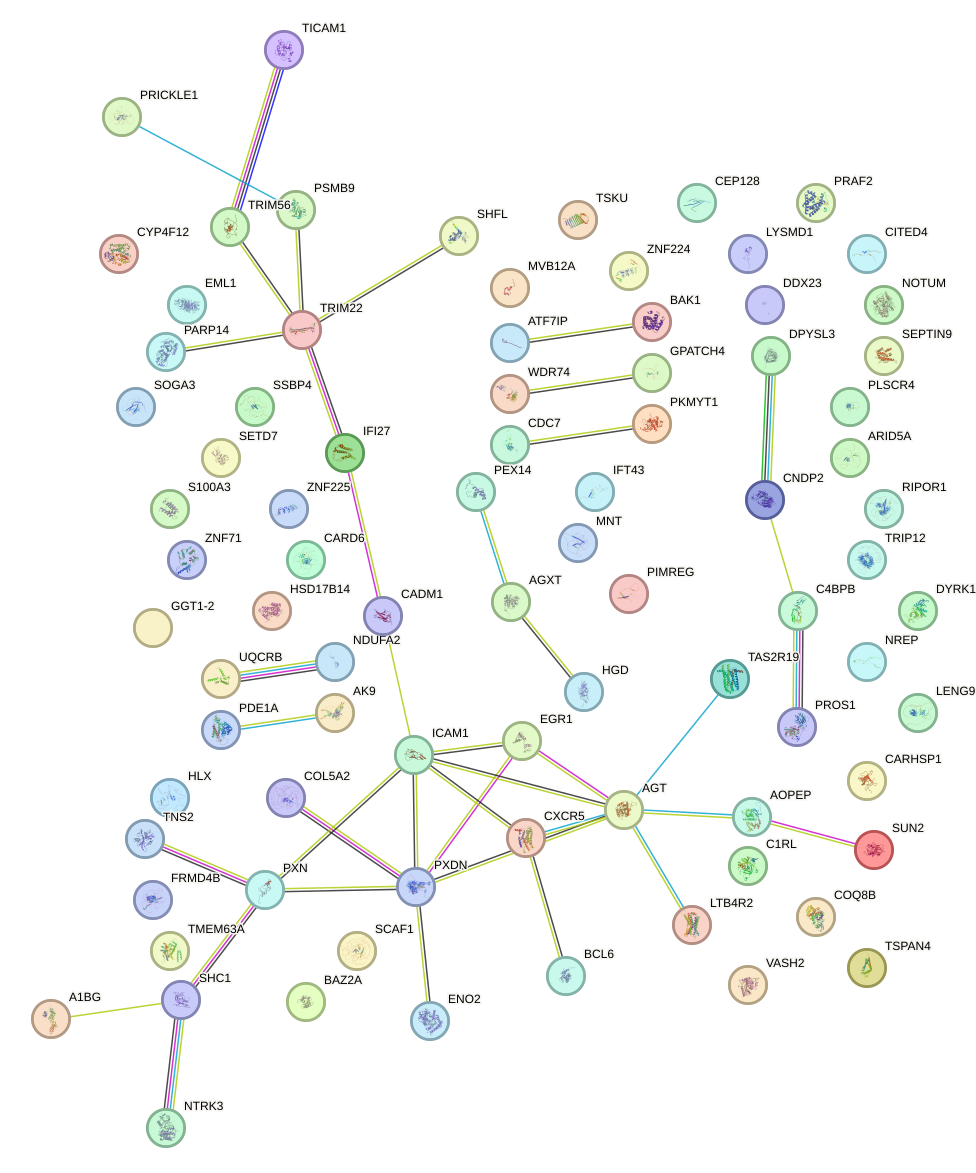
Network of associatons between targets according to the STRING database.
First level regulatory network of STAT1_STAT3_BCL6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.1 | 0.2 | GO:0048597 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.1 | 0.2 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.1 | 0.2 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:1903410 | lysine import(GO:0034226) L-lysine import(GO:0061461) L-lysine import into cell(GO:1903410) |
| 0.0 | 0.2 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 0.0 | 0.1 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.1 | GO:0061567 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.0 | 0.1 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.1 | GO:0019056 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.0 | 0.1 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.0 | 0.1 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.2 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.3 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0003095 | pressure natriuresis(GO:0003095) vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.2 | GO:0045359 | positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.0 | 0.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.2 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.2 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.0 | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome(GO:0098968) |
| 0.0 | 0.0 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 | 0.0 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.0 | 0.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.0 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.0 | 0.0 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.0 | 0.0 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.1 | GO:0019075 | virus maturation(GO:0019075) |
| 0.0 | 0.3 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.1 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.0 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.0 | 0.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.0 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.1 | 0.3 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.3 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.7 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.1 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.0 | 0.1 | GO:0001632 | leukotriene B4 receptor activity(GO:0001632) leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.0 | 0.1 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.0 | 0.1 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 0.1 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.0 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.1 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.0 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.0 | 0.2 | GO:0051400 | BH domain binding(GO:0051400) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |