Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
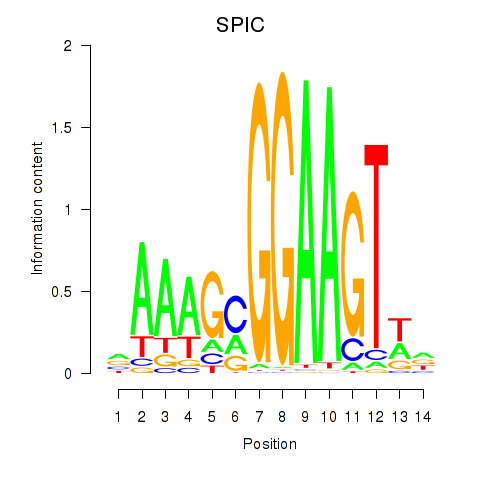
Results for SPIC
Z-value: 0.28
Transcription factors associated with SPIC
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SPIC
|
ENSG00000166211.6 | Spi-C transcription factor |
Activity profile of SPIC motif
Sorted Z-values of SPIC motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_110888103 | 0.12 |
ENST00000426440.1
ENST00000228825.7 |
ARPC3
|
actin related protein 2/3 complex, subunit 3, 21kDa |
| chr7_+_108210012 | 0.11 |
ENST00000249356.3
|
DNAJB9
|
DnaJ (Hsp40) homolog, subfamily B, member 9 |
| chr1_-_149859466 | 0.09 |
ENST00000331128.3
|
HIST2H2AB
|
histone cluster 2, H2ab |
| chr17_+_28443819 | 0.08 |
ENST00000479218.2
|
NSRP1
|
nuclear speckle splicing regulatory protein 1 |
| chr17_-_46690839 | 0.08 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr3_-_121379739 | 0.08 |
ENST00000428394.2
ENST00000314583.3 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr1_-_43855444 | 0.07 |
ENST00000372455.4
|
MED8
|
mediator complex subunit 8 |
| chr21_+_17566643 | 0.07 |
ENST00000419952.1
ENST00000445461.2 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr12_+_13061894 | 0.07 |
ENST00000540125.1
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A |
| chr1_-_33116128 | 0.07 |
ENST00000436661.1
ENST00000373501.2 ENST00000341885.5 ENST00000468695.1 |
ZBTB8OS
|
zinc finger and BTB domain containing 8 opposite strand |
| chr3_+_28390637 | 0.07 |
ENST00000420223.1
ENST00000383768.2 |
ZCWPW2
|
zinc finger, CW type with PWWP domain 2 |
| chr4_+_184427235 | 0.06 |
ENST00000412117.1
ENST00000434682.2 |
ING2
|
inhibitor of growth family, member 2 |
| chr7_-_108209897 | 0.06 |
ENST00000313516.5
|
THAP5
|
THAP domain containing 5 |
| chr3_-_47324008 | 0.06 |
ENST00000425853.1
|
KIF9
|
kinesin family member 9 |
| chr1_+_112016414 | 0.06 |
ENST00000343534.5
ENST00000369718.3 |
C1orf162
|
chromosome 1 open reading frame 162 |
| chr1_-_235324772 | 0.05 |
ENST00000408888.3
|
RBM34
|
RNA binding motif protein 34 |
| chr4_-_103682071 | 0.05 |
ENST00000505239.1
|
MANBA
|
mannosidase, beta A, lysosomal |
| chr18_-_61089665 | 0.05 |
ENST00000238497.5
|
VPS4B
|
vacuolar protein sorting 4 homolog B (S. cerevisiae) |
| chr1_-_43638168 | 0.05 |
ENST00000431635.2
|
EBNA1BP2
|
EBNA1 binding protein 2 |
| chrX_+_100646190 | 0.05 |
ENST00000471855.1
|
RPL36A
|
ribosomal protein L36a |
| chr10_+_35416090 | 0.05 |
ENST00000354759.3
|
CREM
|
cAMP responsive element modulator |
| chr1_-_157789850 | 0.05 |
ENST00000491942.1
ENST00000358292.3 ENST00000368176.3 |
FCRL1
|
Fc receptor-like 1 |
| chr9_-_19380196 | 0.05 |
ENST00000315377.4
ENST00000380384.1 ENST00000380381.3 ENST00000380394.4 |
RPS6
|
ribosomal protein S6 |
| chr7_-_23145288 | 0.05 |
ENST00000419813.1
|
KLHL7-AS1
|
KLHL7 antisense RNA 1 (head to head) |
| chr1_-_235324530 | 0.05 |
ENST00000447801.1
ENST00000366606.3 ENST00000429912.1 |
RBM34
|
RNA binding motif protein 34 |
| chr6_+_26183958 | 0.05 |
ENST00000356530.3
|
HIST1H2BE
|
histone cluster 1, H2be |
| chr19_-_52307357 | 0.05 |
ENST00000594900.1
|
FPR1
|
formyl peptide receptor 1 |
| chr4_-_74853897 | 0.05 |
ENST00000296028.3
|
PPBP
|
pro-platelet basic protein (chemokine (C-X-C motif) ligand 7) |
| chr3_-_47324242 | 0.05 |
ENST00000456548.1
ENST00000432493.1 ENST00000335044.2 ENST00000444589.2 |
KIF9
|
kinesin family member 9 |
| chr1_+_154947126 | 0.05 |
ENST00000368439.1
|
CKS1B
|
CDC28 protein kinase regulatory subunit 1B |
| chr3_-_28390120 | 0.05 |
ENST00000334100.6
|
AZI2
|
5-azacytidine induced 2 |
| chr2_-_40006289 | 0.05 |
ENST00000260619.6
ENST00000454352.2 |
THUMPD2
|
THUMP domain containing 2 |
| chr12_+_6881678 | 0.05 |
ENST00000441671.2
ENST00000203629.2 |
LAG3
|
lymphocyte-activation gene 3 |
| chr10_+_18948311 | 0.05 |
ENST00000377275.3
|
ARL5B
|
ADP-ribosylation factor-like 5B |
| chr6_+_139349817 | 0.05 |
ENST00000367660.3
|
ABRACL
|
ABRA C-terminal like |
| chr3_+_23847432 | 0.05 |
ENST00000346855.3
|
UBE2E1
|
ubiquitin-conjugating enzyme E2E 1 |
| chrX_-_135056106 | 0.05 |
ENST00000433339.2
|
MMGT1
|
membrane magnesium transporter 1 |
| chr15_-_64455404 | 0.05 |
ENST00000300026.3
|
PPIB
|
peptidylprolyl isomerase B (cyclophilin B) |
| chr1_+_202830876 | 0.05 |
ENST00000456105.2
|
RP11-480I12.7
|
RP11-480I12.7 |
| chr17_+_18684563 | 0.05 |
ENST00000476139.1
|
TVP23B
|
trans-golgi network vesicle protein 23 homolog B (S. cerevisiae) |
| chr16_+_81812863 | 0.05 |
ENST00000359376.3
|
PLCG2
|
phospholipase C, gamma 2 (phosphatidylinositol-specific) |
| chr7_+_74188309 | 0.05 |
ENST00000289473.4
ENST00000433458.1 |
NCF1
|
neutrophil cytosolic factor 1 |
| chr7_+_99006550 | 0.05 |
ENST00000222969.5
|
BUD31
|
BUD31 homolog (S. cerevisiae) |
| chrX_-_51239425 | 0.05 |
ENST00000375992.3
|
NUDT11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11 |
| chr2_-_122494487 | 0.05 |
ENST00000451734.1
ENST00000285814.4 |
MKI67IP
|
nucleolar protein interacting with the FHA domain of MKI67 |
| chrX_-_20159934 | 0.05 |
ENST00000379593.1
ENST00000379607.5 |
EIF1AX
|
eukaryotic translation initiation factor 1A, X-linked |
| chr12_-_81331460 | 0.05 |
ENST00000549417.1
|
LIN7A
|
lin-7 homolog A (C. elegans) |
| chr4_+_74606223 | 0.04 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr5_+_112227311 | 0.04 |
ENST00000391338.1
|
ZRSR1
|
zinc finger (CCCH type), RNA-binding motif and serine/arginine rich 1 |
| chr11_-_32816156 | 0.04 |
ENST00000531481.1
ENST00000335185.5 |
CCDC73
|
coiled-coil domain containing 73 |
| chr12_-_110883346 | 0.04 |
ENST00000547365.1
|
ARPC3
|
actin related protein 2/3 complex, subunit 3, 21kDa |
| chr2_+_85822857 | 0.04 |
ENST00000306368.4
ENST00000414390.1 ENST00000456023.1 |
RNF181
|
ring finger protein 181 |
| chr19_+_45542295 | 0.04 |
ENST00000221455.3
ENST00000391953.4 ENST00000588936.1 |
CLASRP
|
CLK4-associating serine/arginine rich protein |
| chr3_+_130613226 | 0.04 |
ENST00000509662.1
ENST00000328560.8 ENST00000428331.2 ENST00000359644.3 ENST00000422190.2 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr16_-_29465668 | 0.04 |
ENST00000569622.1
|
RP11-345J4.5
|
BolA-like protein 2 |
| chr4_-_80994210 | 0.04 |
ENST00000403729.2
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr11_-_68671264 | 0.04 |
ENST00000362034.2
|
MRPL21
|
mitochondrial ribosomal protein L21 |
| chr3_+_130613001 | 0.04 |
ENST00000504948.1
ENST00000513801.1 ENST00000505072.1 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr2_+_113342011 | 0.04 |
ENST00000324913.5
|
CHCHD5
|
coiled-coil-helix-coiled-coil-helix domain containing 5 |
| chr10_-_112064665 | 0.04 |
ENST00000369603.5
|
SMNDC1
|
survival motor neuron domain containing 1 |
| chr3_-_179322416 | 0.04 |
ENST00000259038.2
|
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr4_-_37687991 | 0.04 |
ENST00000314117.4
ENST00000454158.2 |
RELL1
|
RELT-like 1 |
| chr2_+_217363793 | 0.04 |
ENST00000456586.1
ENST00000598925.1 ENST00000427280.2 |
RPL37A
|
ribosomal protein L37a |
| chr19_-_5838768 | 0.04 |
ENST00000527106.1
ENST00000531199.1 ENST00000529165.1 |
FUT6
|
fucosyltransferase 6 (alpha (1,3) fucosyltransferase) |
| chr17_-_7835228 | 0.04 |
ENST00000303731.4
ENST00000571947.1 ENST00000540486.1 ENST00000572656.1 |
TRAPPC1
|
trafficking protein particle complex 1 |
| chr11_-_116658695 | 0.04 |
ENST00000429220.1
ENST00000444935.1 |
ZNF259
|
zinc finger protein 259 |
| chr17_-_37009882 | 0.04 |
ENST00000378096.3
ENST00000394332.1 ENST00000394333.1 ENST00000577407.1 ENST00000479035.2 |
RPL23
|
ribosomal protein L23 |
| chr11_+_118889456 | 0.04 |
ENST00000528230.1
ENST00000525303.1 ENST00000434101.2 ENST00000359005.4 ENST00000533058.1 |
TRAPPC4
|
trafficking protein particle complex 4 |
| chr2_-_175351744 | 0.04 |
ENST00000295500.4
ENST00000392552.2 ENST00000392551.2 |
GPR155
|
G protein-coupled receptor 155 |
| chr11_+_94706804 | 0.04 |
ENST00000335080.5
|
KDM4D
|
lysine (K)-specific demethylase 4D |
| chr16_+_66613351 | 0.04 |
ENST00000379486.2
ENST00000268595.2 |
CMTM2
|
CKLF-like MARVEL transmembrane domain containing 2 |
| chr14_-_23284675 | 0.04 |
ENST00000555959.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr2_-_242211359 | 0.04 |
ENST00000444092.1
|
HDLBP
|
high density lipoprotein binding protein |
| chr11_+_127140956 | 0.04 |
ENST00000608214.1
|
RP11-480C22.1
|
RP11-480C22.1 |
| chr12_-_120663792 | 0.04 |
ENST00000546532.1
ENST00000548912.1 |
PXN
|
paxillin |
| chr11_-_68671244 | 0.04 |
ENST00000567045.1
ENST00000450904.2 |
MRPL21
|
mitochondrial ribosomal protein L21 |
| chr2_+_85822839 | 0.04 |
ENST00000441634.1
|
RNF181
|
ring finger protein 181 |
| chr15_-_80263506 | 0.04 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr5_+_82373379 | 0.04 |
ENST00000396027.4
ENST00000511817.1 |
XRCC4
|
X-ray repair complementing defective repair in Chinese hamster cells 4 |
| chr6_-_33239712 | 0.04 |
ENST00000436044.2
|
VPS52
|
vacuolar protein sorting 52 homolog (S. cerevisiae) |
| chr5_+_59726565 | 0.04 |
ENST00000412930.2
|
FKSG52
|
FKSG52 |
| chr5_-_60458179 | 0.04 |
ENST00000507416.1
ENST00000339020.3 |
SMIM15
|
small integral membrane protein 15 |
| chr7_+_99006232 | 0.04 |
ENST00000403633.2
|
BUD31
|
BUD31 homolog (S. cerevisiae) |
| chr12_+_57854274 | 0.04 |
ENST00000528432.1
|
GLI1
|
GLI family zinc finger 1 |
| chr5_-_40798263 | 0.04 |
ENST00000296800.4
ENST00000397128.2 |
PRKAA1
|
protein kinase, AMP-activated, alpha 1 catalytic subunit |
| chr5_-_140013224 | 0.04 |
ENST00000498971.2
|
CD14
|
CD14 molecule |
| chr5_+_121297650 | 0.04 |
ENST00000339397.4
|
SRFBP1
|
serum response factor binding protein 1 |
| chr12_+_120907622 | 0.04 |
ENST00000392509.2
ENST00000549649.1 ENST00000548342.1 |
DYNLL1
|
dynein, light chain, LC8-type 1 |
| chr2_-_40006357 | 0.04 |
ENST00000505747.1
|
THUMPD2
|
THUMP domain containing 2 |
| chr16_+_81528948 | 0.04 |
ENST00000539778.2
|
CMIP
|
c-Maf inducing protein |
| chr11_-_116658758 | 0.04 |
ENST00000227322.3
|
ZNF259
|
zinc finger protein 259 |
| chr18_-_33077556 | 0.04 |
ENST00000589273.1
ENST00000586489.1 |
INO80C
|
INO80 complex subunit C |
| chr18_-_74839891 | 0.04 |
ENST00000581878.1
|
MBP
|
myelin basic protein |
| chr11_-_71639613 | 0.03 |
ENST00000528184.1
ENST00000528511.2 |
RP11-849H4.2
|
Putative short transient receptor potential channel 2-like protein |
| chr2_-_264024 | 0.03 |
ENST00000403712.2
ENST00000356150.5 ENST00000405430.1 |
SH3YL1
|
SH3 and SYLF domain containing 1 |
| chr22_-_37880543 | 0.03 |
ENST00000442496.1
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr5_-_180235755 | 0.03 |
ENST00000502678.1
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr5_+_82373317 | 0.03 |
ENST00000282268.3
ENST00000338635.6 |
XRCC4
|
X-ray repair complementing defective repair in Chinese hamster cells 4 |
| chr1_+_150245099 | 0.03 |
ENST00000369099.3
|
C1orf54
|
chromosome 1 open reading frame 54 |
| chr1_-_212588157 | 0.03 |
ENST00000261455.4
ENST00000535273.1 |
TMEM206
|
transmembrane protein 206 |
| chr20_-_33264886 | 0.03 |
ENST00000217446.3
ENST00000452740.2 ENST00000374820.2 |
PIGU
|
phosphatidylinositol glycan anchor biosynthesis, class U |
| chr17_+_42925270 | 0.03 |
ENST00000253410.2
ENST00000587021.1 |
HIGD1B
|
HIG1 hypoxia inducible domain family, member 1B |
| chr8_+_104426942 | 0.03 |
ENST00000297579.5
|
DCAF13
|
DDB1 and CUL4 associated factor 13 |
| chr19_-_39826639 | 0.03 |
ENST00000602185.1
ENST00000598034.1 ENST00000601387.1 ENST00000595636.1 ENST00000253054.8 ENST00000594700.1 ENST00000597595.1 |
GMFG
|
glia maturation factor, gamma |
| chr2_-_114300213 | 0.03 |
ENST00000446595.1
ENST00000416105.1 ENST00000450636.1 ENST00000416758.1 |
RP11-395L14.4
|
RP11-395L14.4 |
| chr7_-_108210048 | 0.03 |
ENST00000415914.3
ENST00000438865.1 |
THAP5
|
THAP domain containing 5 |
| chrX_-_135056216 | 0.03 |
ENST00000305963.2
|
MMGT1
|
membrane magnesium transporter 1 |
| chr3_-_47324079 | 0.03 |
ENST00000352910.4
|
KIF9
|
kinesin family member 9 |
| chr8_+_58890917 | 0.03 |
ENST00000522992.1
|
RP11-1112C15.1
|
RP11-1112C15.1 |
| chr19_-_51920835 | 0.03 |
ENST00000442846.3
ENST00000530476.1 |
SIGLEC10
|
sialic acid binding Ig-like lectin 10 |
| chr3_-_47324060 | 0.03 |
ENST00000452770.2
|
KIF9
|
kinesin family member 9 |
| chr15_-_90358048 | 0.03 |
ENST00000300060.6
ENST00000560137.1 |
ANPEP
|
alanyl (membrane) aminopeptidase |
| chr12_-_120907374 | 0.03 |
ENST00000550458.1
|
SRSF9
|
serine/arginine-rich splicing factor 9 |
| chr19_+_55987998 | 0.03 |
ENST00000591164.1
|
ZNF628
|
zinc finger protein 628 |
| chr1_-_43637915 | 0.03 |
ENST00000236051.2
|
EBNA1BP2
|
EBNA1 binding protein 2 |
| chr12_-_57113333 | 0.03 |
ENST00000550920.1
|
NACA
|
nascent polypeptide-associated complex alpha subunit |
| chr1_-_43855479 | 0.03 |
ENST00000290663.6
ENST00000372457.4 |
MED8
|
mediator complex subunit 8 |
| chr2_+_163175394 | 0.03 |
ENST00000446271.1
ENST00000429691.2 |
GCA
|
grancalcin, EF-hand calcium binding protein |
| chr19_-_51875894 | 0.03 |
ENST00000600427.1
ENST00000595217.1 ENST00000221978.5 |
NKG7
|
natural killer cell group 7 sequence |
| chr17_+_37356555 | 0.03 |
ENST00000579374.1
|
RPL19
|
ribosomal protein L19 |
| chr1_-_109618566 | 0.03 |
ENST00000338366.5
|
TAF13
|
TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa |
| chr11_-_85780853 | 0.03 |
ENST00000531930.1
ENST00000528398.1 |
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr18_-_34408693 | 0.03 |
ENST00000587382.1
ENST00000589049.1 ENST00000587129.1 |
TPGS2
|
tubulin polyglutamylase complex subunit 2 |
| chr12_-_8693539 | 0.03 |
ENST00000299663.3
|
CLEC4E
|
C-type lectin domain family 4, member E |
| chr19_-_51920952 | 0.03 |
ENST00000356298.5
ENST00000339313.5 ENST00000529627.1 ENST00000439889.2 ENST00000353836.5 ENST00000432469.2 |
SIGLEC10
|
sialic acid binding Ig-like lectin 10 |
| chrX_-_11129229 | 0.03 |
ENST00000608176.1
ENST00000433747.2 ENST00000608576.1 ENST00000608916.1 |
RP11-120D5.1
|
RP11-120D5.1 |
| chr6_+_31554826 | 0.03 |
ENST00000376089.2
ENST00000396112.2 |
LST1
|
leukocyte specific transcript 1 |
| chr14_+_77924204 | 0.03 |
ENST00000555133.1
|
AHSA1
|
AHA1, activator of heat shock 90kDa protein ATPase homolog 1 (yeast) |
| chr2_+_202125219 | 0.03 |
ENST00000323492.7
|
CASP8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr19_+_58694396 | 0.03 |
ENST00000326804.4
ENST00000345813.3 ENST00000424679.2 |
ZNF274
|
zinc finger protein 274 |
| chr11_-_7698453 | 0.03 |
ENST00000524608.1
|
CYB5R2
|
cytochrome b5 reductase 2 |
| chr11_-_36531774 | 0.03 |
ENST00000348124.5
ENST00000526995.1 |
TRAF6
|
TNF receptor-associated factor 6, E3 ubiquitin protein ligase |
| chr14_-_89021077 | 0.03 |
ENST00000556564.1
|
PTPN21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr19_+_45542773 | 0.03 |
ENST00000544944.2
|
CLASRP
|
CLK4-associating serine/arginine rich protein |
| chr2_-_231084617 | 0.03 |
ENST00000409815.2
|
SP110
|
SP110 nuclear body protein |
| chr19_-_10450328 | 0.03 |
ENST00000160262.5
|
ICAM3
|
intercellular adhesion molecule 3 |
| chrX_-_71497077 | 0.03 |
ENST00000373626.3
|
RPS4X
|
ribosomal protein S4, X-linked |
| chr3_-_28390298 | 0.03 |
ENST00000457172.1
|
AZI2
|
5-azacytidine induced 2 |
| chr4_-_47916543 | 0.03 |
ENST00000507489.1
|
NFXL1
|
nuclear transcription factor, X-box binding-like 1 |
| chr3_+_149530836 | 0.03 |
ENST00000466478.1
ENST00000491086.1 ENST00000467977.1 |
RNF13
|
ring finger protein 13 |
| chr7_+_23145884 | 0.03 |
ENST00000409689.1
ENST00000410047.1 |
KLHL7
|
kelch-like family member 7 |
| chr5_+_115177178 | 0.03 |
ENST00000316788.7
|
AP3S1
|
adaptor-related protein complex 3, sigma 1 subunit |
| chr1_+_179335101 | 0.03 |
ENST00000508285.1
ENST00000511889.1 |
AXDND1
|
axonemal dynein light chain domain containing 1 |
| chr3_+_148847371 | 0.03 |
ENST00000296051.2
ENST00000460120.1 |
HPS3
|
Hermansky-Pudlak syndrome 3 |
| chr1_+_40505891 | 0.03 |
ENST00000372797.3
ENST00000372802.1 ENST00000449311.1 |
CAP1
|
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr5_+_68530668 | 0.03 |
ENST00000506563.1
|
CDK7
|
cyclin-dependent kinase 7 |
| chr20_+_44486246 | 0.03 |
ENST00000255152.2
ENST00000454862.2 |
ZSWIM3
|
zinc finger, SWIM-type containing 3 |
| chr14_-_81687197 | 0.03 |
ENST00000553612.1
|
GTF2A1
|
general transcription factor IIA, 1, 19/37kDa |
| chr2_-_220119280 | 0.03 |
ENST00000392088.2
|
TUBA4A
|
tubulin, alpha 4a |
| chr7_-_102252589 | 0.03 |
ENST00000520042.1
|
RASA4
|
RAS p21 protein activator 4 |
| chr19_-_10450287 | 0.03 |
ENST00000589261.1
ENST00000590569.1 ENST00000589580.1 ENST00000589249.1 |
ICAM3
|
intercellular adhesion molecule 3 |
| chr11_+_118889142 | 0.03 |
ENST00000533632.1
|
TRAPPC4
|
trafficking protein particle complex 4 |
| chr1_+_207262881 | 0.03 |
ENST00000451804.2
|
C4BPB
|
complement component 4 binding protein, beta |
| chr18_+_6256746 | 0.03 |
ENST00000578427.1
|
RP11-760N9.1
|
RP11-760N9.1 |
| chr17_+_41005283 | 0.03 |
ENST00000592999.1
|
AOC3
|
amine oxidase, copper containing 3 |
| chr9_-_95087838 | 0.03 |
ENST00000442668.2
ENST00000421075.2 ENST00000536624.1 |
NOL8
|
nucleolar protein 8 |
| chr1_-_6420737 | 0.03 |
ENST00000541130.1
ENST00000377845.3 |
ACOT7
|
acyl-CoA thioesterase 7 |
| chr3_+_179322573 | 0.03 |
ENST00000493866.1
ENST00000472629.1 ENST00000482604.1 |
NDUFB5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa |
| chr5_+_10250328 | 0.03 |
ENST00000515390.1
|
CCT5
|
chaperonin containing TCP1, subunit 5 (epsilon) |
| chr7_-_151330218 | 0.03 |
ENST00000476632.1
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr15_-_90233866 | 0.03 |
ENST00000561257.1
|
PEX11A
|
peroxisomal biogenesis factor 11 alpha |
| chr4_-_99064387 | 0.03 |
ENST00000295268.3
|
STPG2
|
sperm-tail PG-rich repeat containing 2 |
| chr4_-_80994619 | 0.03 |
ENST00000404191.1
|
ANTXR2
|
anthrax toxin receptor 2 |
| chrX_-_107018969 | 0.03 |
ENST00000372383.4
|
TSC22D3
|
TSC22 domain family, member 3 |
| chr11_+_102188272 | 0.03 |
ENST00000532808.1
|
BIRC3
|
baculoviral IAP repeat containing 3 |
| chr1_-_154155595 | 0.03 |
ENST00000328159.4
ENST00000368531.2 ENST00000323144.7 ENST00000368533.3 ENST00000341372.3 |
TPM3
|
tropomyosin 3 |
| chr8_+_117950422 | 0.03 |
ENST00000378279.3
|
AARD
|
alanine and arginine rich domain containing protein |
| chr2_+_48667983 | 0.03 |
ENST00000449090.2
|
PPP1R21
|
protein phosphatase 1, regulatory subunit 21 |
| chr10_-_119806085 | 0.03 |
ENST00000355624.3
|
RAB11FIP2
|
RAB11 family interacting protein 2 (class I) |
| chr14_+_63671105 | 0.03 |
ENST00000316754.3
|
RHOJ
|
ras homolog family member J |
| chr2_-_32264850 | 0.03 |
ENST00000295066.3
ENST00000342166.5 |
DPY30
|
dpy-30 homolog (C. elegans) |
| chr22_+_18593446 | 0.03 |
ENST00000316027.6
|
TUBA8
|
tubulin, alpha 8 |
| chr1_+_150245177 | 0.03 |
ENST00000369098.3
|
C1orf54
|
chromosome 1 open reading frame 54 |
| chr17_-_18161870 | 0.03 |
ENST00000579294.1
ENST00000545457.2 ENST00000379450.4 ENST00000578558.1 |
FLII
|
flightless I homolog (Drosophila) |
| chr5_+_94890840 | 0.03 |
ENST00000504763.1
|
ARSK
|
arylsulfatase family, member K |
| chr12_+_21654714 | 0.03 |
ENST00000542038.1
ENST00000540141.1 ENST00000229314.5 |
GOLT1B
|
golgi transport 1B |
| chr14_+_65878650 | 0.03 |
ENST00000555559.1
|
FUT8
|
fucosyltransferase 8 (alpha (1,6) fucosyltransferase) |
| chr12_+_16500037 | 0.03 |
ENST00000536371.1
ENST00000010404.2 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr19_+_32896697 | 0.03 |
ENST00000586987.1
|
DPY19L3
|
dpy-19-like 3 (C. elegans) |
| chr4_+_57333756 | 0.03 |
ENST00000510663.1
ENST00000504757.1 |
SRP72
|
signal recognition particle 72kDa |
| chr3_+_130612803 | 0.03 |
ENST00000510168.1
ENST00000508532.1 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr1_-_169337176 | 0.03 |
ENST00000472647.1
ENST00000367811.3 |
NME7
|
NME/NM23 family member 7 |
| chr5_+_96079240 | 0.03 |
ENST00000515663.1
|
CAST
|
calpastatin |
| chr3_+_122296465 | 0.03 |
ENST00000483793.1
|
PARP15
|
poly (ADP-ribose) polymerase family, member 15 |
| chr2_+_109065634 | 0.03 |
ENST00000409821.1
|
GCC2
|
GRIP and coiled-coil domain containing 2 |
| chr6_+_36973406 | 0.03 |
ENST00000274963.8
|
FGD2
|
FYVE, RhoGEF and PH domain containing 2 |
| chrX_-_63615297 | 0.03 |
ENST00000374852.3
ENST00000453546.1 |
MTMR8
|
myotubularin related protein 8 |
| chr6_+_170863421 | 0.03 |
ENST00000392092.2
ENST00000540980.1 ENST00000230354.6 |
TBP
|
TATA box binding protein |
| chr17_+_37356586 | 0.03 |
ENST00000579260.1
ENST00000582193.1 |
RPL19
|
ribosomal protein L19 |
| chr12_-_77272765 | 0.03 |
ENST00000547435.1
ENST00000552330.1 ENST00000546966.1 ENST00000311083.5 |
CSRP2
|
cysteine and glycine-rich protein 2 |
| chr11_-_71639670 | 0.03 |
ENST00000533047.1
ENST00000529844.1 |
RP11-849H4.2
|
Putative short transient receptor potential channel 2-like protein |
| chr19_-_50464338 | 0.03 |
ENST00000426971.2
ENST00000447370.2 |
SIGLEC11
|
sialic acid binding Ig-like lectin 11 |
| chr21_+_34638656 | 0.03 |
ENST00000290200.2
|
IL10RB
|
interleukin 10 receptor, beta |
| chrX_-_47863348 | 0.03 |
ENST00000376943.3
ENST00000396965.1 ENST00000305127.6 |
ZNF182
|
zinc finger protein 182 |
| chr6_+_14117872 | 0.03 |
ENST00000379153.3
|
CD83
|
CD83 molecule |
| chr8_+_41348173 | 0.02 |
ENST00000357743.4
|
GOLGA7
|
golgin A7 |
| chr5_+_134181625 | 0.02 |
ENST00000394976.3
|
C5orf24
|
chromosome 5 open reading frame 24 |
| chr1_+_104068562 | 0.02 |
ENST00000423855.2
|
RNPC3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr5_-_150460914 | 0.02 |
ENST00000389378.2
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr4_-_112993808 | 0.02 |
ENST00000511219.1
|
RP11-269F21.3
|
RP11-269F21.3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SPIC
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0048611 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.1 | GO:1903722 | negative regulation of exosomal secretion(GO:1903542) regulation of centriole elongation(GO:1903722) |
| 0.0 | 0.0 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) |
| 0.0 | 0.0 | GO:0006679 | glucosylceramide biosynthetic process(GO:0006679) |
| 0.0 | 0.1 | GO:0038123 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.0 | 0.1 | GO:0052056 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.0 | 0.0 | GO:2000417 | negative regulation of eosinophil migration(GO:2000417) |
| 0.0 | 0.1 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.0 | GO:1902954 | regulation of early endosome to recycling endosome transport(GO:1902954) |
| 0.0 | 0.1 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.0 | 0.0 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 0.0 | 0.1 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.1 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.0 | 0.0 | GO:0002290 | gamma-delta T cell activation involved in immune response(GO:0002290) negative regulation of interferon-beta secretion(GO:0035548) regulation of gamma-delta T cell activation involved in immune response(GO:2001191) positive regulation of gamma-delta T cell activation involved in immune response(GO:2001193) |
| 0.0 | 0.1 | GO:1901908 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.0 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.0 | 0.0 | GO:0001300 | chronological cell aging(GO:0001300) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.1 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.0 | GO:0072563 | endothelial microparticle(GO:0072563) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.0 | GO:0047322 | [hydroxymethylglutaryl-CoA reductase (NADPH)] kinase activity(GO:0047322) [acetyl-CoA carboxylase] kinase activity(GO:0050405) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.0 | GO:0032396 | HLA-A specific inhibitory MHC class I receptor activity(GO:0030107) HLA-B specific inhibitory MHC class I receptor activity(GO:0030109) inhibitory MHC class I receptor activity(GO:0032396) |
| 0.0 | 0.1 | GO:0052840 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.0 | 0.1 | GO:0001132 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |