Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
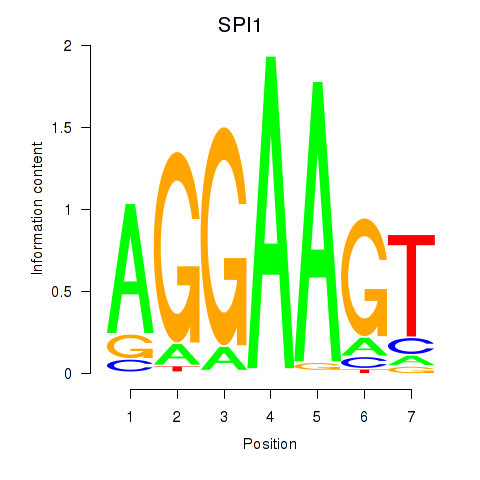
Results for SPI1
Z-value: 0.60
Transcription factors associated with SPI1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SPI1
|
ENSG00000066336.7 | Spi-1 proto-oncogene |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SPI1 | hg19_v2_chr11_-_47400062_47400077 | -0.86 | 1.4e-01 | Click! |
Activity profile of SPI1 motif
Sorted Z-values of SPI1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_75532373 | 0.37 |
ENST00000595757.1
|
AC022400.2
|
Uncharacterized protein; cDNA FLJ44715 fis, clone BRACE3021430 |
| chr11_-_3862206 | 0.35 |
ENST00000351018.4
|
RHOG
|
ras homolog family member G |
| chr2_-_220408260 | 0.32 |
ENST00000373891.2
|
CHPF
|
chondroitin polymerizing factor |
| chr9_+_2017063 | 0.31 |
ENST00000457226.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr15_-_64385981 | 0.24 |
ENST00000557835.1
ENST00000380290.3 ENST00000559950.1 |
FAM96A
|
family with sequence similarity 96, member A |
| chr8_+_22438009 | 0.23 |
ENST00000409417.1
|
PDLIM2
|
PDZ and LIM domain 2 (mystique) |
| chr14_-_23284703 | 0.21 |
ENST00000555911.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr1_+_16083123 | 0.20 |
ENST00000510393.1
ENST00000430076.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr11_-_3862059 | 0.20 |
ENST00000396978.1
|
RHOG
|
ras homolog family member G |
| chr14_-_23285069 | 0.20 |
ENST00000554758.1
ENST00000397528.4 |
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr17_-_79269067 | 0.19 |
ENST00000288439.5
ENST00000374759.3 |
SLC38A10
|
solute carrier family 38, member 10 |
| chr1_+_16083098 | 0.19 |
ENST00000496928.2
ENST00000508310.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr17_-_19651598 | 0.19 |
ENST00000570414.1
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr16_-_30394143 | 0.18 |
ENST00000321367.3
ENST00000571393.1 |
SEPT1
|
septin 1 |
| chr5_+_180650271 | 0.18 |
ENST00000351937.5
ENST00000315073.5 |
TRIM41
|
tripartite motif containing 41 |
| chr1_+_38273419 | 0.18 |
ENST00000468084.1
|
C1orf122
|
chromosome 1 open reading frame 122 |
| chr12_+_94071129 | 0.18 |
ENST00000552983.1
ENST00000332896.3 ENST00000552033.1 ENST00000548483.1 |
CRADD
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr6_-_34524093 | 0.18 |
ENST00000544425.1
|
SPDEF
|
SAM pointed domain containing ETS transcription factor |
| chr2_-_153573965 | 0.17 |
ENST00000448428.1
|
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr9_+_140172200 | 0.17 |
ENST00000357503.2
|
TOR4A
|
torsin family 4, member A |
| chr1_-_151254362 | 0.17 |
ENST00000447795.2
|
RP11-126K1.2
|
Uncharacterized protein |
| chr7_-_75241096 | 0.17 |
ENST00000420909.1
|
HIP1
|
huntingtin interacting protein 1 |
| chr19_+_39421556 | 0.16 |
ENST00000407800.2
ENST00000402029.3 |
MRPS12
|
mitochondrial ribosomal protein S12 |
| chr5_-_140053152 | 0.16 |
ENST00000542735.1
|
DND1
|
DND microRNA-mediated repression inhibitor 1 |
| chr14_+_102276192 | 0.16 |
ENST00000557714.1
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr6_+_144980954 | 0.16 |
ENST00000367525.3
|
UTRN
|
utrophin |
| chr3_+_49027771 | 0.16 |
ENST00000475629.1
ENST00000444213.1 |
P4HTM
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) |
| chr8_+_22437965 | 0.16 |
ENST00000409141.1
ENST00000265810.4 |
PDLIM2
|
PDZ and LIM domain 2 (mystique) |
| chr17_-_33415837 | 0.16 |
ENST00000414419.2
|
RFFL
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr11_+_394145 | 0.15 |
ENST00000528036.1
|
PKP3
|
plakophilin 3 |
| chr19_-_3557570 | 0.15 |
ENST00000355415.2
|
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr11_-_67120974 | 0.15 |
ENST00000539074.1
ENST00000312419.3 |
POLD4
|
polymerase (DNA-directed), delta 4, accessory subunit |
| chr10_-_126849626 | 0.15 |
ENST00000530884.1
|
CTBP2
|
C-terminal binding protein 2 |
| chr16_+_30205225 | 0.15 |
ENST00000345535.4
ENST00000251303.6 |
SLX1A
|
SLX1 structure-specific endonuclease subunit homolog A (S. cerevisiae) |
| chr6_-_32143828 | 0.15 |
ENST00000412465.2
ENST00000375107.3 |
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr1_-_52520828 | 0.14 |
ENST00000610127.1
|
TXNDC12
|
thioredoxin domain containing 12 (endoplasmic reticulum) |
| chr6_-_33168391 | 0.14 |
ENST00000374685.4
ENST00000413614.2 ENST00000374680.3 |
RXRB
|
retinoid X receptor, beta |
| chr6_+_100054606 | 0.14 |
ENST00000369215.4
|
PRDM13
|
PR domain containing 13 |
| chr6_-_32160622 | 0.14 |
ENST00000487761.1
ENST00000375040.3 |
GPSM3
|
G-protein signaling modulator 3 |
| chr16_-_31085514 | 0.14 |
ENST00000300849.4
|
ZNF668
|
zinc finger protein 668 |
| chr17_-_8093471 | 0.14 |
ENST00000389017.4
|
C17orf59
|
chromosome 17 open reading frame 59 |
| chr16_+_29465822 | 0.14 |
ENST00000330181.5
ENST00000351581.4 |
SLX1B
|
SLX1 structure-specific endonuclease subunit homolog B (S. cerevisiae) |
| chr19_+_41257084 | 0.14 |
ENST00000601393.1
|
SNRPA
|
small nuclear ribonucleoprotein polypeptide A |
| chr14_+_74004051 | 0.14 |
ENST00000557556.1
|
ACOT1
|
acyl-CoA thioesterase 1 |
| chr6_-_33266687 | 0.13 |
ENST00000444031.2
|
RGL2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chr17_+_74261277 | 0.13 |
ENST00000327490.6
|
UBALD2
|
UBA-like domain containing 2 |
| chr1_+_16083154 | 0.13 |
ENST00000375771.1
|
FBLIM1
|
filamin binding LIM protein 1 |
| chr17_+_45908974 | 0.13 |
ENST00000269025.4
|
LRRC46
|
leucine rich repeat containing 46 |
| chr16_+_31085714 | 0.13 |
ENST00000300850.5
ENST00000564189.1 ENST00000428260.1 |
ZNF646
|
zinc finger protein 646 |
| chr14_-_23285011 | 0.13 |
ENST00000397532.3
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr1_+_84767289 | 0.13 |
ENST00000394834.3
ENST00000370669.1 |
SAMD13
|
sterile alpha motif domain containing 13 |
| chr11_-_72433346 | 0.13 |
ENST00000334211.8
|
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr19_+_17530838 | 0.13 |
ENST00000528659.1
ENST00000392702.2 ENST00000529939.1 |
MVB12A
|
multivesicular body subunit 12A |
| chr7_-_5569588 | 0.13 |
ENST00000417101.1
|
ACTB
|
actin, beta |
| chr11_+_64879317 | 0.13 |
ENST00000526809.1
ENST00000279263.7 ENST00000524986.1 ENST00000534371.1 ENST00000540748.1 ENST00000525385.1 ENST00000345348.5 ENST00000531321.1 ENST00000529414.1 ENST00000526085.1 ENST00000530750.1 |
TM7SF2
|
transmembrane 7 superfamily member 2 |
| chr16_+_67261008 | 0.13 |
ENST00000304800.9
ENST00000563953.1 ENST00000565201.1 |
TMEM208
|
transmembrane protein 208 |
| chr16_-_88717482 | 0.13 |
ENST00000261623.3
|
CYBA
|
cytochrome b-245, alpha polypeptide |
| chr3_+_50388126 | 0.12 |
ENST00000425346.1
ENST00000424512.1 ENST00000232508.5 ENST00000418577.1 ENST00000606589.1 |
CYB561D2
XXcos-LUCA11.5
|
cytochrome b561 family, member D2 Uncharacterized protein |
| chr16_-_2205352 | 0.12 |
ENST00000563192.1
|
RP11-304L19.5
|
RP11-304L19.5 |
| chr15_-_55488817 | 0.12 |
ENST00000569386.1
|
RSL24D1
|
ribosomal L24 domain containing 1 |
| chr2_+_239756671 | 0.12 |
ENST00000448943.2
|
TWIST2
|
twist family bHLH transcription factor 2 |
| chr17_-_7760457 | 0.12 |
ENST00000576384.1
|
LSMD1
|
LSM domain containing 1 |
| chr17_+_74261413 | 0.12 |
ENST00000587913.1
|
UBALD2
|
UBA-like domain containing 2 |
| chr9_+_135037334 | 0.12 |
ENST00000393229.3
ENST00000360670.3 ENST00000393228.4 ENST00000372179.3 |
NTNG2
|
netrin G2 |
| chr1_+_198126209 | 0.12 |
ENST00000367383.1
|
NEK7
|
NIMA-related kinase 7 |
| chr17_-_7761256 | 0.12 |
ENST00000575208.1
|
LSMD1
|
LSM domain containing 1 |
| chr20_+_62711482 | 0.12 |
ENST00000336866.2
ENST00000355631.4 |
OPRL1
|
opiate receptor-like 1 |
| chr6_-_154751629 | 0.12 |
ENST00000424998.1
|
CNKSR3
|
CNKSR family member 3 |
| chr6_-_33267101 | 0.12 |
ENST00000497454.1
|
RGL2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chr17_-_4710288 | 0.12 |
ENST00000571067.1
|
RP11-81A22.5
|
RP11-81A22.5 |
| chr17_+_42422662 | 0.12 |
ENST00000593167.1
ENST00000585512.1 ENST00000591740.1 ENST00000592783.1 ENST00000587387.1 ENST00000588237.1 ENST00000589265.1 |
GRN
|
granulin |
| chr1_+_31883048 | 0.11 |
ENST00000536859.1
|
SERINC2
|
serine incorporator 2 |
| chr2_+_219262948 | 0.11 |
ENST00000443891.1
|
CTDSP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 |
| chr4_+_8201091 | 0.11 |
ENST00000382521.3
ENST00000245105.3 ENST00000457650.2 ENST00000539824.1 |
SH3TC1
|
SH3 domain and tetratricopeptide repeats 1 |
| chr11_+_64073699 | 0.11 |
ENST00000405666.1
ENST00000468670.1 |
ESRRA
|
estrogen-related receptor alpha |
| chr9_-_134145880 | 0.11 |
ENST00000372269.3
ENST00000464831.1 |
FAM78A
|
family with sequence similarity 78, member A |
| chr22_+_37678505 | 0.11 |
ENST00000402997.1
ENST00000405206.3 |
CYTH4
|
cytohesin 4 |
| chr20_+_43990576 | 0.11 |
ENST00000372727.1
ENST00000414310.1 |
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr19_+_36236491 | 0.11 |
ENST00000591949.1
|
PSENEN
|
presenilin enhancer gamma secretase subunit |
| chr2_+_138721850 | 0.11 |
ENST00000329366.4
ENST00000280097.3 |
HNMT
|
histamine N-methyltransferase |
| chr6_-_6320875 | 0.11 |
ENST00000451619.1
|
F13A1
|
coagulation factor XIII, A1 polypeptide |
| chr11_-_62358972 | 0.11 |
ENST00000278279.3
|
TUT1
|
terminal uridylyl transferase 1, U6 snRNA-specific |
| chr2_+_220094657 | 0.11 |
ENST00000436226.1
|
ANKZF1
|
ankyrin repeat and zinc finger domain containing 1 |
| chr1_+_221051699 | 0.11 |
ENST00000366903.6
|
HLX
|
H2.0-like homeobox |
| chr11_+_66824346 | 0.11 |
ENST00000532559.1
|
RHOD
|
ras homolog family member D |
| chr4_+_926171 | 0.11 |
ENST00000507319.1
ENST00000264771.4 |
TMEM175
|
transmembrane protein 175 |
| chr11_-_62359027 | 0.11 |
ENST00000494385.1
ENST00000308436.7 |
TUT1
|
terminal uridylyl transferase 1, U6 snRNA-specific |
| chr6_-_34524049 | 0.11 |
ENST00000374037.3
|
SPDEF
|
SAM pointed domain containing ETS transcription factor |
| chr17_-_43138463 | 0.11 |
ENST00000310604.4
|
DCAKD
|
dephospho-CoA kinase domain containing |
| chr11_+_66624527 | 0.11 |
ENST00000393952.3
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr19_+_15218180 | 0.11 |
ENST00000342784.2
ENST00000597977.1 ENST00000600440.1 |
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans) |
| chr16_-_67260901 | 0.11 |
ENST00000341546.3
ENST00000409509.1 ENST00000433915.1 ENST00000454102.2 |
LRRC29
AC040160.1
|
leucine rich repeat containing 29 Uncharacterized protein; cDNA FLJ57407, weakly similar to Mus musculus leucine rich repeat containing 29 (Lrrc29), mRNA |
| chr3_+_181429704 | 0.10 |
ENST00000431565.2
ENST00000325404.1 |
SOX2
|
SRY (sex determining region Y)-box 2 |
| chr4_-_140223614 | 0.10 |
ENST00000394223.1
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr16_-_67260691 | 0.10 |
ENST00000447579.1
ENST00000393992.1 ENST00000424285.1 |
LRRC29
|
leucine rich repeat containing 29 |
| chr19_-_12780211 | 0.10 |
ENST00000597961.1
ENST00000598732.1 ENST00000222190.5 |
CTD-2192J16.24
WDR83OS
|
Uncharacterized protein WD repeat domain 83 opposite strand |
| chr14_-_77495007 | 0.10 |
ENST00000238647.3
|
IRF2BPL
|
interferon regulatory factor 2 binding protein-like |
| chr11_-_118789613 | 0.10 |
ENST00000532899.1
|
BCL9L
|
B-cell CLL/lymphoma 9-like |
| chr8_+_99076750 | 0.10 |
ENST00000545282.1
|
C8orf47
|
chromosome 8 open reading frame 47 |
| chr11_+_45825616 | 0.10 |
ENST00000442528.2
ENST00000456334.1 ENST00000526817.1 |
SLC35C1
|
solute carrier family 35 (GDP-fucose transporter), member C1 |
| chr8_-_144886321 | 0.10 |
ENST00000526832.1
|
SCRIB
|
scribbled planar cell polarity protein |
| chr6_-_113754604 | 0.10 |
ENST00000421737.1
|
RP1-124C6.1
|
RP1-124C6.1 |
| chr4_-_153601136 | 0.10 |
ENST00000504064.1
ENST00000304385.3 |
TMEM154
|
transmembrane protein 154 |
| chr19_+_35739897 | 0.10 |
ENST00000605618.1
ENST00000427250.1 ENST00000601623.1 |
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr4_+_926214 | 0.10 |
ENST00000514453.1
ENST00000515492.1 ENST00000509508.1 ENST00000515740.1 ENST00000508204.1 ENST00000510493.1 ENST00000514546.1 |
TMEM175
|
transmembrane protein 175 |
| chr22_+_27068704 | 0.10 |
ENST00000444388.1
ENST00000450963.1 ENST00000449017.1 |
CTA-211A9.5
|
CTA-211A9.5 |
| chr3_-_178789220 | 0.10 |
ENST00000414084.1
|
ZMAT3
|
zinc finger, matrin-type 3 |
| chr11_+_128563652 | 0.10 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr17_-_6917755 | 0.09 |
ENST00000593646.1
|
AC040977.1
|
Uncharacterized protein |
| chr12_+_53443680 | 0.09 |
ENST00000314250.6
ENST00000451358.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr11_+_47430133 | 0.09 |
ENST00000531974.1
ENST00000531419.1 ENST00000531865.1 ENST00000362021.4 ENST00000354884.4 |
SLC39A13
|
solute carrier family 39 (zinc transporter), member 13 |
| chr11_+_2421718 | 0.09 |
ENST00000380996.5
ENST00000333256.6 ENST00000380992.1 ENST00000437110.1 ENST00000435795.1 |
TSSC4
|
tumor suppressing subtransferable candidate 4 |
| chr21_+_33671264 | 0.09 |
ENST00000339944.4
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr11_-_62389449 | 0.09 |
ENST00000534026.1
|
B3GAT3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chr11_-_62389621 | 0.09 |
ENST00000531383.1
ENST00000265471.5 |
B3GAT3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chr7_+_150759634 | 0.09 |
ENST00000392826.2
ENST00000461735.1 |
SLC4A2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr12_-_118796910 | 0.09 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr11_+_64107663 | 0.09 |
ENST00000356786.5
|
CCDC88B
|
coiled-coil domain containing 88B |
| chr22_+_38004832 | 0.09 |
ENST00000405147.3
ENST00000429218.1 ENST00000325180.8 ENST00000337437.4 |
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr1_-_153917700 | 0.09 |
ENST00000368646.2
|
DENND4B
|
DENN/MADD domain containing 4B |
| chr12_-_58135903 | 0.09 |
ENST00000257897.3
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr11_+_844406 | 0.09 |
ENST00000397404.1
|
TSPAN4
|
tetraspanin 4 |
| chr17_+_75315654 | 0.09 |
ENST00000590595.1
|
SEPT9
|
septin 9 |
| chr2_+_32502952 | 0.09 |
ENST00000238831.4
|
YIPF4
|
Yip1 domain family, member 4 |
| chr12_-_56652111 | 0.09 |
ENST00000267116.7
|
ANKRD52
|
ankyrin repeat domain 52 |
| chr3_-_11623804 | 0.09 |
ENST00000451674.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr2_+_28615669 | 0.09 |
ENST00000379619.1
ENST00000264716.4 |
FOSL2
|
FOS-like antigen 2 |
| chr17_+_38599693 | 0.09 |
ENST00000542955.1
ENST00000269593.4 |
IGFBP4
|
insulin-like growth factor binding protein 4 |
| chr1_-_154909329 | 0.09 |
ENST00000368467.3
|
PMVK
|
phosphomevalonate kinase |
| chr19_+_14551066 | 0.08 |
ENST00000342216.4
|
PKN1
|
protein kinase N1 |
| chr5_-_141060389 | 0.08 |
ENST00000504448.1
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr1_+_151254738 | 0.08 |
ENST00000336715.6
ENST00000324048.5 ENST00000368879.2 |
ZNF687
|
zinc finger protein 687 |
| chr10_+_75532028 | 0.08 |
ENST00000372841.3
ENST00000394790.1 |
FUT11
|
fucosyltransferase 11 (alpha (1,3) fucosyltransferase) |
| chr15_-_55611306 | 0.08 |
ENST00000563262.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr17_+_65027509 | 0.08 |
ENST00000375684.1
|
AC005544.1
|
Uncharacterized protein |
| chr15_-_64386120 | 0.08 |
ENST00000300030.3
|
FAM96A
|
family with sequence similarity 96, member A |
| chr17_-_7760779 | 0.08 |
ENST00000335155.5
ENST00000575071.1 |
LSMD1
|
LSM domain containing 1 |
| chr17_-_37844267 | 0.08 |
ENST00000579146.1
ENST00000378011.4 ENST00000429199.2 ENST00000300658.4 |
PGAP3
|
post-GPI attachment to proteins 3 |
| chr11_+_120081475 | 0.08 |
ENST00000328965.4
|
OAF
|
OAF homolog (Drosophila) |
| chr17_+_49243639 | 0.08 |
ENST00000512737.1
ENST00000503064.1 |
NME1-NME2
|
NME1-NME2 readthrough |
| chr16_-_88717423 | 0.08 |
ENST00000568278.1
ENST00000569359.1 ENST00000567174.1 |
CYBA
|
cytochrome b-245, alpha polypeptide |
| chr9_-_138987115 | 0.08 |
ENST00000277554.2
|
NACC2
|
NACC family member 2, BEN and BTB (POZ) domain containing |
| chr16_+_28875268 | 0.08 |
ENST00000395532.4
|
SH2B1
|
SH2B adaptor protein 1 |
| chr12_+_6493319 | 0.08 |
ENST00000536876.1
|
LTBR
|
lymphotoxin beta receptor (TNFR superfamily, member 3) |
| chr4_-_926069 | 0.08 |
ENST00000314167.4
ENST00000502656.1 |
GAK
|
cyclin G associated kinase |
| chr17_+_2264983 | 0.08 |
ENST00000574650.1
|
SGSM2
|
small G protein signaling modulator 2 |
| chr17_+_4843352 | 0.08 |
ENST00000573404.1
ENST00000576452.1 |
RNF167
|
ring finger protein 167 |
| chr9_+_139557360 | 0.08 |
ENST00000308874.7
ENST00000406555.3 ENST00000492862.2 |
EGFL7
|
EGF-like-domain, multiple 7 |
| chr14_-_68157084 | 0.08 |
ENST00000557564.1
|
RP11-1012A1.4
|
RP11-1012A1.4 |
| chr7_-_100076873 | 0.08 |
ENST00000300181.2
|
TSC22D4
|
TSC22 domain family, member 4 |
| chr3_-_149293990 | 0.08 |
ENST00000472417.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr16_-_29875057 | 0.08 |
ENST00000219789.6
|
CDIPT
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase |
| chr11_-_72432950 | 0.08 |
ENST00000426523.1
ENST00000429686.1 |
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr16_-_30393752 | 0.08 |
ENST00000566517.1
ENST00000605106.1 |
SEPT1
SEPT1
|
septin 1 Uncharacterized protein |
| chr14_+_65170820 | 0.08 |
ENST00000555982.1
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr14_+_65171099 | 0.08 |
ENST00000247226.7
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr17_+_7255208 | 0.08 |
ENST00000333751.3
|
KCTD11
|
potassium channel tetramerization domain containing 11 |
| chr16_+_3162557 | 0.08 |
ENST00000382192.3
ENST00000219091.4 ENST00000444510.2 ENST00000414351.1 |
ZNF205
|
zinc finger protein 205 |
| chr15_-_52971544 | 0.08 |
ENST00000566768.1
ENST00000561543.1 |
FAM214A
|
family with sequence similarity 214, member A |
| chr14_+_50159813 | 0.08 |
ENST00000359332.2
ENST00000553274.1 ENST00000557128.1 |
KLHDC1
|
kelch domain containing 1 |
| chr16_+_28875126 | 0.08 |
ENST00000359285.5
ENST00000538342.1 |
SH2B1
|
SH2B adaptor protein 1 |
| chr6_-_33266492 | 0.07 |
ENST00000425946.1
|
RGL2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chr12_-_92539614 | 0.07 |
ENST00000256015.3
|
BTG1
|
B-cell translocation gene 1, anti-proliferative |
| chr17_-_40134339 | 0.07 |
ENST00000587727.1
|
DNAJC7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr14_+_24563510 | 0.07 |
ENST00000545054.2
ENST00000561286.1 ENST00000558096.1 |
PCK2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr16_-_28506840 | 0.07 |
ENST00000569430.1
|
CLN3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr16_-_68404908 | 0.07 |
ENST00000574662.1
|
SMPD3
|
sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II) |
| chr14_+_23340822 | 0.07 |
ENST00000359591.4
|
LRP10
|
low density lipoprotein receptor-related protein 10 |
| chr19_-_4559814 | 0.07 |
ENST00000586582.1
|
SEMA6B
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B |
| chr14_-_50999373 | 0.07 |
ENST00000554273.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr17_-_43510282 | 0.07 |
ENST00000290470.3
|
ARHGAP27
|
Rho GTPase activating protein 27 |
| chr3_-_187455680 | 0.07 |
ENST00000438077.1
|
BCL6
|
B-cell CLL/lymphoma 6 |
| chr18_-_60985914 | 0.07 |
ENST00000589955.1
|
BCL2
|
B-cell CLL/lymphoma 2 |
| chr3_-_50360192 | 0.07 |
ENST00000442581.1
ENST00000447092.1 ENST00000357750.4 |
HYAL2
|
hyaluronoglucosaminidase 2 |
| chr4_+_103422499 | 0.07 |
ENST00000511926.1
ENST00000507079.1 |
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| chr19_-_12792704 | 0.07 |
ENST00000210060.7
|
DHPS
|
deoxyhypusine synthase |
| chr22_-_39150947 | 0.07 |
ENST00000411587.2
ENST00000420859.1 ENST00000452294.1 ENST00000456894.1 |
SUN2
|
Sad1 and UNC84 domain containing 2 |
| chr19_+_48248779 | 0.07 |
ENST00000246802.5
|
GLTSCR2
|
glioma tumor suppressor candidate region gene 2 |
| chr3_+_9774164 | 0.07 |
ENST00000426583.1
|
BRPF1
|
bromodomain and PHD finger containing, 1 |
| chr17_-_26697304 | 0.07 |
ENST00000536498.1
|
VTN
|
vitronectin |
| chr7_-_143579973 | 0.07 |
ENST00000460532.1
ENST00000491908.1 |
FAM115A
|
family with sequence similarity 115, member A |
| chrX_-_49056635 | 0.07 |
ENST00000472598.1
ENST00000538567.1 ENST00000479808.1 ENST00000263233.4 |
SYP
|
synaptophysin |
| chr6_+_33168597 | 0.07 |
ENST00000374675.3
|
SLC39A7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr2_+_220462560 | 0.07 |
ENST00000456909.1
ENST00000295641.10 |
STK11IP
|
serine/threonine kinase 11 interacting protein |
| chr19_+_42387228 | 0.07 |
ENST00000354532.3
ENST00000599846.1 ENST00000347545.4 |
ARHGEF1
|
Rho guanine nucleotide exchange factor (GEF) 1 |
| chr11_-_6502534 | 0.07 |
ENST00000254584.2
ENST00000525235.1 ENST00000445086.2 |
ARFIP2
|
ADP-ribosylation factor interacting protein 2 |
| chr6_-_32145861 | 0.07 |
ENST00000336984.6
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr19_+_36119929 | 0.07 |
ENST00000588161.1
ENST00000262633.4 ENST00000592202.1 ENST00000586618.1 |
RBM42
|
RNA binding motif protein 42 |
| chr16_+_20911174 | 0.07 |
ENST00000568663.1
|
LYRM1
|
LYR motif containing 1 |
| chr8_-_121457608 | 0.07 |
ENST00000306185.3
|
MRPL13
|
mitochondrial ribosomal protein L13 |
| chr18_+_21594384 | 0.07 |
ENST00000584250.1
|
TTC39C
|
tetratricopeptide repeat domain 39C |
| chr17_+_4843679 | 0.07 |
ENST00000576229.1
|
RNF167
|
ring finger protein 167 |
| chr5_+_174151536 | 0.07 |
ENST00000239243.6
ENST00000507785.1 |
MSX2
|
msh homeobox 2 |
| chr2_-_183291741 | 0.07 |
ENST00000351439.5
ENST00000409365.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr16_-_67597789 | 0.07 |
ENST00000605277.1
|
CTD-2012K14.6
|
CTD-2012K14.6 |
| chr4_+_109571800 | 0.07 |
ENST00000512478.2
|
OSTC
|
oligosaccharyltransferase complex subunit (non-catalytic) |
| chr17_+_8191815 | 0.07 |
ENST00000226105.6
ENST00000407006.4 ENST00000580434.1 ENST00000439238.3 |
RANGRF
|
RAN guanine nucleotide release factor |
| chr1_-_157108266 | 0.07 |
ENST00000326786.4
|
ETV3
|
ets variant 3 |
| chr14_-_23451467 | 0.07 |
ENST00000555074.1
ENST00000361265.4 |
RP11-298I3.5
AJUBA
|
RP11-298I3.5 ajuba LIM protein |
| chr20_+_43374421 | 0.07 |
ENST00000372861.3
|
KCNK15
|
potassium channel, subfamily K, member 15 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SPI1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.1 | 0.2 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.0 | 0.1 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.3 | GO:0032485 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.3 | GO:0060481 | lung goblet cell differentiation(GO:0060480) lobar bronchus epithelium development(GO:0060481) |
| 0.0 | 0.4 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.0 | 0.2 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.1 | GO:0002352 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 0.2 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.0 | 0.1 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.0 | 0.1 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.0 | 0.1 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.0 | 0.2 | GO:0036079 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.5 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.1 | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome(GO:0098968) |
| 0.0 | 0.1 | GO:0046203 | spermidine catabolic process(GO:0046203) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.1 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.0 | 0.1 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.0 | 0.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.1 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.2 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0032848 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.0 | 0.1 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.1 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.1 | GO:0019075 | virus maturation(GO:0019075) |
| 0.0 | 0.1 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.3 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:0033029 | regulation of neutrophil apoptotic process(GO:0033029) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.1 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.0 | 0.1 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.1 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.1 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 0.0 | GO:0034445 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.0 | 0.1 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.1 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.0 | 0.1 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.0 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.0 | 0.0 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.0 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 0.0 | 0.1 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.1 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.0 | 0.1 | GO:0019087 | transformation of host cell by virus(GO:0019087) renal water absorption(GO:0070295) |
| 0.0 | 0.5 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.0 | 0.1 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.1 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.1 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.3 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.1 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.6 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.1 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.0 | 0.1 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.1 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.0 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.0 | 0.0 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.1 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.1 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.0 | 0.1 | GO:0070236 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:0033087 | negative regulation of immature T cell proliferation(GO:0033087) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 0.0 | 0.2 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.1 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 0.0 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.1 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.0 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.0 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.0 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.1 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.2 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.1 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.2 | GO:0046930 | pore complex(GO:0046930) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 0.2 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.1 | GO:0034038 | deoxyhypusine synthase activity(GO:0034038) |
| 0.0 | 0.1 | GO:0031781 | melanocortin receptor binding(GO:0031779) corticotropin hormone receptor binding(GO:0031780) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) type 5 melanocortin receptor binding(GO:0031783) type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.1 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.0 | 0.2 | GO:0036080 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.0 | 0.3 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 0.0 | 0.1 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 0.1 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.0 | 0.1 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.2 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.1 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.1 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.3 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.4 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.3 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.1 | GO:0033906 | protein tyrosine kinase inhibitor activity(GO:0030292) hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.3 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.3 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.0 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.1 | GO:0004974 | leukotriene B4 receptor activity(GO:0001632) leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.0 | GO:0005115 | receptor tyrosine kinase-like orphan receptor binding(GO:0005115) chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.0 | 0.0 | GO:0086040 | sodium:proton antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086040) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.0 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.0 | 0.1 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.3 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.1 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.0 | 0.1 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.2 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |