Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
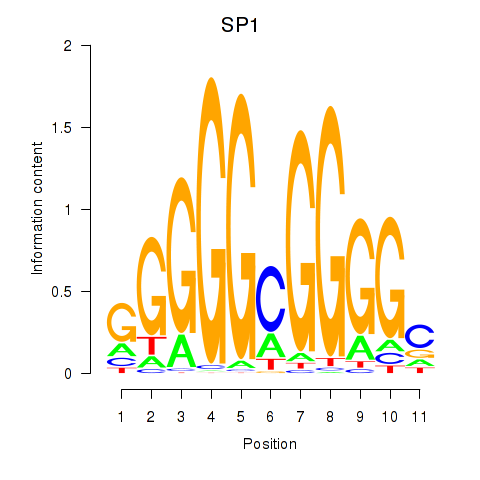
Results for SP1
Z-value: 0.17
Transcription factors associated with SP1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SP1
|
ENSG00000185591.5 | Sp1 transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SP1 | hg19_v2_chr12_+_53773944_53773993 | -0.75 | 2.5e-01 | Click! |
Activity profile of SP1 motif
Sorted Z-values of SP1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_15001430 | 0.11 |
ENST00000407572.1
|
MEIG1
|
meiosis/spermiogenesis associated 1 |
| chr19_+_55795493 | 0.11 |
ENST00000309383.1
|
BRSK1
|
BR serine/threonine kinase 1 |
| chr13_-_52027134 | 0.10 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr8_-_99837856 | 0.09 |
ENST00000518165.1
ENST00000419617.2 |
STK3
|
serine/threonine kinase 3 |
| chr11_+_64009072 | 0.09 |
ENST00000535135.1
ENST00000394540.3 |
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr11_+_64008525 | 0.09 |
ENST00000449942.2
|
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr6_-_168397757 | 0.08 |
ENST00000456585.1
ENST00000414364.1 |
KIF25-AS1
|
KIF25 antisense RNA 1 |
| chr14_+_21566980 | 0.08 |
ENST00000418511.2
ENST00000554329.2 |
TMEM253
|
transmembrane protein 253 |
| chr19_-_18314799 | 0.08 |
ENST00000481914.2
|
RAB3A
|
RAB3A, member RAS oncogene family |
| chr16_-_31021717 | 0.08 |
ENST00000565419.1
|
STX1B
|
syntaxin 1B |
| chr11_+_65686728 | 0.07 |
ENST00000312515.2
ENST00000525501.1 |
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr9_+_129987488 | 0.07 |
ENST00000446764.1
|
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3 |
| chr11_+_65686802 | 0.07 |
ENST00000376991.2
|
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr17_-_72968837 | 0.07 |
ENST00000581676.1
|
HID1
|
HID1 domain containing |
| chr1_+_211432700 | 0.07 |
ENST00000452621.2
|
RCOR3
|
REST corepressor 3 |
| chr17_-_58469329 | 0.06 |
ENST00000393003.3
|
USP32
|
ubiquitin specific peptidase 32 |
| chr11_+_64008443 | 0.06 |
ENST00000309366.4
|
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr2_+_99758161 | 0.06 |
ENST00000409684.1
|
C2ORF15
|
Uncharacterized protein C2orf15 |
| chr1_+_211432775 | 0.06 |
ENST00000419091.2
|
RCOR3
|
REST corepressor 3 |
| chr11_-_34937858 | 0.06 |
ENST00000278359.5
|
APIP
|
APAF1 interacting protein |
| chr19_+_1261106 | 0.06 |
ENST00000588411.1
|
CIRBP
|
cold inducible RNA binding protein |
| chr1_-_85156417 | 0.06 |
ENST00000422026.1
|
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr10_-_112678904 | 0.06 |
ENST00000423273.1
ENST00000436562.1 ENST00000447005.1 ENST00000454061.1 |
BBIP1
|
BBSome interacting protein 1 |
| chr3_-_113415441 | 0.06 |
ENST00000491165.1
ENST00000316407.4 |
KIAA2018
|
KIAA2018 |
| chr6_-_41863098 | 0.06 |
ENST00000373006.1
|
USP49
|
ubiquitin specific peptidase 49 |
| chr2_-_45162783 | 0.06 |
ENST00000432125.2
|
RP11-89K21.1
|
RP11-89K21.1 |
| chr1_-_23670752 | 0.06 |
ENST00000302271.6
ENST00000426846.2 ENST00000427764.2 ENST00000606561.1 ENST00000374616.3 |
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R |
| chr16_+_90086002 | 0.06 |
ENST00000563936.1
ENST00000536122.1 ENST00000561675.1 |
GAS8
|
growth arrest-specific 8 |
| chr12_-_104531785 | 0.06 |
ENST00000551727.1
|
NFYB
|
nuclear transcription factor Y, beta |
| chr4_-_102268708 | 0.06 |
ENST00000525819.1
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr7_+_151038850 | 0.06 |
ENST00000355851.4
ENST00000566856.1 ENST00000470229.1 |
NUB1
|
negative regulator of ubiquitin-like proteins 1 |
| chr3_-_57113281 | 0.06 |
ENST00000468466.1
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3 |
| chr2_-_232328867 | 0.06 |
ENST00000453992.1
ENST00000417652.1 ENST00000454824.1 |
NCL
|
nucleolin |
| chrX_-_38186775 | 0.06 |
ENST00000339363.3
ENST00000309513.3 ENST00000338898.3 ENST00000342811.3 ENST00000378505.2 |
RPGR
|
retinitis pigmentosa GTPase regulator |
| chr8_-_29120604 | 0.06 |
ENST00000521515.1
|
KIF13B
|
kinesin family member 13B |
| chr17_-_58469687 | 0.06 |
ENST00000590133.1
|
USP32
|
ubiquitin specific peptidase 32 |
| chr6_-_110500826 | 0.06 |
ENST00000265601.3
ENST00000447287.1 ENST00000444391.1 |
WASF1
|
WAS protein family, member 1 |
| chr22_-_38966123 | 0.06 |
ENST00000439567.1
|
DMC1
|
DNA meiotic recombinase 1 |
| chr16_-_89268070 | 0.05 |
ENST00000562855.2
|
SLC22A31
|
solute carrier family 22, member 31 |
| chr10_+_76586348 | 0.05 |
ENST00000372724.1
ENST00000287239.4 ENST00000372714.1 |
KAT6B
|
K(lysine) acetyltransferase 6B |
| chr11_+_65686952 | 0.05 |
ENST00000527119.1
|
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr18_+_77867177 | 0.05 |
ENST00000560752.1
|
ADNP2
|
ADNP homeobox 2 |
| chr9_-_34523027 | 0.05 |
ENST00000399775.2
|
ENHO
|
energy homeostasis associated |
| chr17_-_15466742 | 0.05 |
ENST00000584811.1
ENST00000419890.2 |
TVP23C
|
trans-golgi network vesicle protein 23 homolog C (S. cerevisiae) |
| chr8_+_26149274 | 0.05 |
ENST00000522535.1
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha |
| chr6_-_86353510 | 0.05 |
ENST00000444272.1
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr20_+_56884752 | 0.05 |
ENST00000244040.3
|
RAB22A
|
RAB22A, member RAS oncogene family |
| chr2_+_121493717 | 0.05 |
ENST00000418323.1
|
GLI2
|
GLI family zinc finger 2 |
| chrX_-_14891150 | 0.05 |
ENST00000452869.1
ENST00000398334.1 ENST00000324138.3 |
FANCB
|
Fanconi anemia, complementation group B |
| chr8_-_124286735 | 0.05 |
ENST00000395571.3
|
ZHX1
|
zinc fingers and homeoboxes 1 |
| chr8_-_92053042 | 0.05 |
ENST00000520014.1
|
TMEM55A
|
transmembrane protein 55A |
| chr5_-_176738883 | 0.05 |
ENST00000513169.1
ENST00000423571.2 ENST00000502529.1 ENST00000427908.2 |
MXD3
|
MAX dimerization protein 3 |
| chr8_+_120885949 | 0.05 |
ENST00000523492.1
ENST00000286234.5 |
DEPTOR
|
DEP domain containing MTOR-interacting protein |
| chr6_-_111136299 | 0.05 |
ENST00000457688.1
|
CDK19
|
cyclin-dependent kinase 19 |
| chr16_-_54963026 | 0.05 |
ENST00000560208.1
ENST00000557792.1 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr11_-_64512469 | 0.05 |
ENST00000377485.1
|
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr3_-_176915036 | 0.05 |
ENST00000427349.1
ENST00000352800.5 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr19_+_30302805 | 0.05 |
ENST00000262643.3
ENST00000575243.1 ENST00000357943.5 |
CCNE1
|
cyclin E1 |
| chr4_-_819880 | 0.05 |
ENST00000505203.1
|
CPLX1
|
complexin 1 |
| chr6_-_4135693 | 0.05 |
ENST00000495548.1
ENST00000380125.2 ENST00000465828.1 |
ECI2
|
enoyl-CoA delta isomerase 2 |
| chr11_-_46142615 | 0.05 |
ENST00000529734.1
ENST00000323180.6 |
PHF21A
|
PHD finger protein 21A |
| chr2_+_24272576 | 0.05 |
ENST00000380986.4
ENST00000452109.1 |
FKBP1B
|
FK506 binding protein 1B, 12.6 kDa |
| chr12_+_1100423 | 0.05 |
ENST00000592048.1
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr13_-_96329048 | 0.05 |
ENST00000606011.1
ENST00000499499.2 |
DNAJC3-AS1
|
DNAJC3 antisense RNA 1 (head to head) |
| chr17_-_27277615 | 0.05 |
ENST00000583747.1
ENST00000584236.1 |
PHF12
|
PHD finger protein 12 |
| chr11_-_96123022 | 0.05 |
ENST00000542662.1
|
CCDC82
|
coiled-coil domain containing 82 |
| chr17_-_48943706 | 0.05 |
ENST00000499247.2
|
TOB1
|
transducer of ERBB2, 1 |
| chr3_+_52444651 | 0.05 |
ENST00000327906.3
|
PHF7
|
PHD finger protein 7 |
| chr10_-_18948208 | 0.05 |
ENST00000607346.1
|
ARL5B-AS1
|
ARL5B antisense RNA 1 |
| chr11_+_65687158 | 0.05 |
ENST00000532933.1
|
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr15_+_63481668 | 0.05 |
ENST00000321437.4
ENST00000559006.1 ENST00000448330.2 |
RAB8B
|
RAB8B, member RAS oncogene family |
| chr11_-_34938039 | 0.05 |
ENST00000395787.3
|
APIP
|
APAF1 interacting protein |
| chr15_+_68346501 | 0.05 |
ENST00000249636.6
|
PIAS1
|
protein inhibitor of activated STAT, 1 |
| chr8_+_110552046 | 0.05 |
ENST00000529931.1
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9 |
| chr11_-_61196858 | 0.05 |
ENST00000413184.2
|
CPSF7
|
cleavage and polyadenylation specific factor 7, 59kDa |
| chr3_+_170075436 | 0.05 |
ENST00000476188.1
ENST00000259119.4 ENST00000426052.2 |
SKIL
|
SKI-like oncogene |
| chr2_+_136289030 | 0.05 |
ENST00000409478.1
ENST00000264160.4 ENST00000329971.3 ENST00000438014.1 |
R3HDM1
|
R3H domain containing 1 |
| chrX_-_38186811 | 0.05 |
ENST00000318842.7
|
RPGR
|
retinitis pigmentosa GTPase regulator |
| chr1_-_23670813 | 0.05 |
ENST00000374612.1
|
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R |
| chr5_-_141257954 | 0.05 |
ENST00000456271.1
ENST00000394536.3 ENST00000503492.1 ENST00000287008.3 |
PCDH1
|
protocadherin 1 |
| chr5_+_40679584 | 0.05 |
ENST00000302472.3
|
PTGER4
|
prostaglandin E receptor 4 (subtype EP4) |
| chr6_-_4135825 | 0.05 |
ENST00000380118.3
ENST00000413766.2 ENST00000361538.2 |
ECI2
|
enoyl-CoA delta isomerase 2 |
| chr17_-_42276574 | 0.05 |
ENST00000589805.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr19_-_47164386 | 0.05 |
ENST00000391916.2
ENST00000410105.2 |
DACT3
|
dishevelled-binding antagonist of beta-catenin 3 |
| chr12_-_57824561 | 0.05 |
ENST00000448732.1
|
R3HDM2
|
R3H domain containing 2 |
| chr11_-_77531752 | 0.05 |
ENST00000440064.2
ENST00000528095.1 |
RSF1
|
remodeling and spacing factor 1 |
| chr7_-_105925367 | 0.05 |
ENST00000354289.4
|
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chr7_-_155089251 | 0.04 |
ENST00000609974.1
|
AC144652.1
|
AC144652.1 |
| chr12_-_30907862 | 0.04 |
ENST00000541765.1
ENST00000537108.1 |
CAPRIN2
|
caprin family member 2 |
| chr12_+_69004805 | 0.04 |
ENST00000541216.1
|
RAP1B
|
RAP1B, member of RAS oncogene family |
| chr7_+_151038785 | 0.04 |
ENST00000413040.2
ENST00000568733.1 |
NUB1
|
negative regulator of ubiquitin-like proteins 1 |
| chr2_+_46926326 | 0.04 |
ENST00000394861.2
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr3_-_52002194 | 0.04 |
ENST00000466412.1
|
PCBP4
|
poly(rC) binding protein 4 |
| chr4_-_83350580 | 0.04 |
ENST00000349655.4
ENST00000602300.1 |
HNRNPDL
|
heterogeneous nuclear ribonucleoprotein D-like |
| chrX_+_69674943 | 0.04 |
ENST00000542398.1
|
DLG3
|
discs, large homolog 3 (Drosophila) |
| chr12_+_69004736 | 0.04 |
ENST00000545720.2
|
RAP1B
|
RAP1B, member of RAS oncogene family |
| chr19_+_11457232 | 0.04 |
ENST00000587531.1
|
CCDC159
|
coiled-coil domain containing 159 |
| chr2_-_37193606 | 0.04 |
ENST00000379213.2
ENST00000263918.4 |
STRN
|
striatin, calmodulin binding protein |
| chr17_-_4710288 | 0.04 |
ENST00000571067.1
|
RP11-81A22.5
|
RP11-81A22.5 |
| chr10_+_86088381 | 0.04 |
ENST00000224756.8
ENST00000372088.2 |
CCSER2
|
coiled-coil serine-rich protein 2 |
| chr3_+_135684515 | 0.04 |
ENST00000264977.3
ENST00000490467.1 |
PPP2R3A
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr3_+_195413160 | 0.04 |
ENST00000599448.1
|
LINC00969
|
long intergenic non-protein coding RNA 969 |
| chr16_-_54962704 | 0.04 |
ENST00000502066.2
ENST00000560912.1 ENST00000558952.1 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr10_+_114709999 | 0.04 |
ENST00000355995.4
ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr21_+_45875354 | 0.04 |
ENST00000291592.4
|
LRRC3
|
leucine rich repeat containing 3 |
| chr11_-_8892464 | 0.04 |
ENST00000527347.1
ENST00000526241.1 ENST00000526126.1 ENST00000530938.1 ENST00000526057.1 |
ST5
|
suppression of tumorigenicity 5 |
| chr11_-_95523500 | 0.04 |
ENST00000540054.1
|
FAM76B
|
family with sequence similarity 76, member B |
| chr10_-_104001231 | 0.04 |
ENST00000370002.3
|
PITX3
|
paired-like homeodomain 3 |
| chr6_-_18155285 | 0.04 |
ENST00000309983.4
|
TPMT
|
thiopurine S-methyltransferase |
| chr6_+_12012170 | 0.04 |
ENST00000487103.1
|
HIVEP1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr8_-_17104099 | 0.04 |
ENST00000524358.1
|
CNOT7
|
CCR4-NOT transcription complex, subunit 7 |
| chr16_-_46865286 | 0.04 |
ENST00000285697.4
|
C16orf87
|
chromosome 16 open reading frame 87 |
| chr5_-_118324200 | 0.04 |
ENST00000515439.3
ENST00000510708.1 |
DTWD2
|
DTW domain containing 2 |
| chr19_+_3224700 | 0.04 |
ENST00000292672.2
ENST00000541430.2 |
CELF5
|
CUGBP, Elav-like family member 5 |
| chr20_-_62462566 | 0.04 |
ENST00000245663.4
ENST00000302995.2 |
ZBTB46
|
zinc finger and BTB domain containing 46 |
| chr16_+_56691838 | 0.04 |
ENST00000394501.2
|
MT1F
|
metallothionein 1F |
| chr1_-_26232951 | 0.04 |
ENST00000426559.2
ENST00000455785.2 |
STMN1
|
stathmin 1 |
| chr2_+_24272543 | 0.04 |
ENST00000380991.4
|
FKBP1B
|
FK506 binding protein 1B, 12.6 kDa |
| chr19_-_55658687 | 0.04 |
ENST00000593046.1
|
TNNT1
|
troponin T type 1 (skeletal, slow) |
| chr19_+_42773371 | 0.04 |
ENST00000571942.2
|
CIC
|
capicua transcriptional repressor |
| chr16_-_49315731 | 0.04 |
ENST00000219197.6
|
CBLN1
|
cerebellin 1 precursor |
| chr6_+_138188351 | 0.04 |
ENST00000421450.1
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr6_-_110501126 | 0.04 |
ENST00000368938.1
|
WASF1
|
WAS protein family, member 1 |
| chr14_-_21994525 | 0.04 |
ENST00000538754.1
|
SALL2
|
spalt-like transcription factor 2 |
| chrX_-_19905577 | 0.04 |
ENST00000379697.3
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr3_+_133292759 | 0.04 |
ENST00000431519.2
|
CDV3
|
CDV3 homolog (mouse) |
| chr11_-_69490135 | 0.04 |
ENST00000542341.1
|
ORAOV1
|
oral cancer overexpressed 1 |
| chr2_-_230786378 | 0.04 |
ENST00000430954.1
|
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr6_-_131384347 | 0.04 |
ENST00000530481.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr12_+_112451222 | 0.04 |
ENST00000552052.1
|
ERP29
|
endoplasmic reticulum protein 29 |
| chr2_-_219433014 | 0.04 |
ENST00000418019.1
ENST00000454775.1 ENST00000338465.5 ENST00000415516.1 ENST00000258399.3 |
USP37
|
ubiquitin specific peptidase 37 |
| chr20_-_40247133 | 0.04 |
ENST00000373233.3
ENST00000309279.7 |
CHD6
|
chromodomain helicase DNA binding protein 6 |
| chr10_+_111767720 | 0.04 |
ENST00000356080.4
ENST00000277900.8 |
ADD3
|
adducin 3 (gamma) |
| chr1_-_26232522 | 0.04 |
ENST00000399728.1
|
STMN1
|
stathmin 1 |
| chr15_+_80351977 | 0.04 |
ENST00000559157.1
ENST00000561012.1 ENST00000564367.1 ENST00000558494.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr15_-_83316711 | 0.04 |
ENST00000568128.1
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr12_+_49761147 | 0.04 |
ENST00000549298.1
|
SPATS2
|
spermatogenesis associated, serine-rich 2 |
| chr1_+_120839005 | 0.04 |
ENST00000369390.3
ENST00000452190.1 |
FAM72B
|
family with sequence similarity 72, member B |
| chr19_-_51568324 | 0.04 |
ENST00000595547.1
ENST00000335422.3 ENST00000595793.1 ENST00000596955.1 |
KLK13
|
kallikrein-related peptidase 13 |
| chr10_+_70883908 | 0.04 |
ENST00000263559.6
ENST00000395098.1 ENST00000546041.1 ENST00000541711.1 |
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe) |
| chr9_+_110045418 | 0.04 |
ENST00000419616.1
|
RAD23B
|
RAD23 homolog B (S. cerevisiae) |
| chr2_-_176033066 | 0.04 |
ENST00000437522.1
|
ATF2
|
activating transcription factor 2 |
| chr2_+_42396574 | 0.04 |
ENST00000401738.3
|
EML4
|
echinoderm microtubule associated protein like 4 |
| chr10_-_12084770 | 0.04 |
ENST00000357604.5
|
UPF2
|
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr7_-_140179146 | 0.04 |
ENST00000437223.2
|
MKRN1
|
makorin ring finger protein 1 |
| chr4_-_56412713 | 0.04 |
ENST00000435527.2
|
CLOCK
|
clock circadian regulator |
| chr1_-_143913143 | 0.04 |
ENST00000400889.1
|
FAM72D
|
family with sequence similarity 72, member D |
| chr17_+_29815013 | 0.04 |
ENST00000394744.2
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II) |
| chr10_+_49514698 | 0.04 |
ENST00000432379.1
ENST00000429041.1 ENST00000374189.1 |
MAPK8
|
mitogen-activated protein kinase 8 |
| chr1_-_23670817 | 0.04 |
ENST00000478691.1
|
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R |
| chr1_+_156308403 | 0.04 |
ENST00000481479.1
ENST00000368252.1 ENST00000466306.1 ENST00000368251.1 |
TSACC
|
TSSK6 activating co-chaperone |
| chr1_-_109584716 | 0.04 |
ENST00000531337.1
ENST00000529074.1 ENST00000369965.4 |
WDR47
|
WD repeat domain 47 |
| chr1_-_156675535 | 0.04 |
ENST00000368221.1
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr17_-_7197881 | 0.04 |
ENST00000007699.5
|
YBX2
|
Y box binding protein 2 |
| chr9_-_27573651 | 0.04 |
ENST00000379995.1
ENST00000379997.3 |
C9orf72
|
chromosome 9 open reading frame 72 |
| chr4_-_129209221 | 0.04 |
ENST00000512483.1
|
PGRMC2
|
progesterone receptor membrane component 2 |
| chr7_-_4923315 | 0.04 |
ENST00000399583.3
|
RADIL
|
Ras association and DIL domains |
| chrX_-_37706815 | 0.04 |
ENST00000378578.4
|
DYNLT3
|
dynein, light chain, Tctex-type 3 |
| chr21_-_43916296 | 0.04 |
ENST00000398352.3
|
RSPH1
|
radial spoke head 1 homolog (Chlamydomonas) |
| chr11_+_117103333 | 0.04 |
ENST00000534428.1
|
RNF214
|
ring finger protein 214 |
| chrX_-_101186981 | 0.04 |
ENST00000458570.1
|
ZMAT1
|
zinc finger, matrin-type 1 |
| chr1_+_112939121 | 0.04 |
ENST00000441739.1
|
CTTNBP2NL
|
CTTNBP2 N-terminal like |
| chr22_+_29469100 | 0.04 |
ENST00000327813.5
ENST00000407188.1 |
KREMEN1
|
kringle containing transmembrane protein 1 |
| chr14_+_60716159 | 0.04 |
ENST00000325658.3
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr16_+_87425914 | 0.04 |
ENST00000565788.1
|
MAP1LC3B
|
microtubule-associated protein 1 light chain 3 beta |
| chr10_+_121485588 | 0.04 |
ENST00000361976.2
ENST00000369083.3 |
INPP5F
|
inositol polyphosphate-5-phosphatase F |
| chr15_+_44084040 | 0.04 |
ENST00000249786.4
|
SERF2
|
small EDRK-rich factor 2 |
| chr19_-_49622348 | 0.04 |
ENST00000408991.2
|
C19orf73
|
chromosome 19 open reading frame 73 |
| chr1_-_54411240 | 0.04 |
ENST00000371378.2
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr11_-_77531858 | 0.04 |
ENST00000360355.2
|
RSF1
|
remodeling and spacing factor 1 |
| chr7_-_99774945 | 0.04 |
ENST00000292377.2
|
GPC2
|
glypican 2 |
| chr17_-_27038765 | 0.04 |
ENST00000581289.1
ENST00000301039.2 |
PROCA1
|
protein interacting with cyclin A1 |
| chr17_+_42081914 | 0.04 |
ENST00000293404.3
ENST00000589767.1 |
NAGS
|
N-acetylglutamate synthase |
| chrX_+_14891598 | 0.04 |
ENST00000497603.2
|
MOSPD2
|
motile sperm domain containing 2 |
| chr12_-_30907822 | 0.04 |
ENST00000540436.1
|
CAPRIN2
|
caprin family member 2 |
| chr6_+_138188378 | 0.04 |
ENST00000420009.1
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr6_-_109761707 | 0.04 |
ENST00000520723.1
ENST00000518648.1 ENST00000417394.2 |
PPIL6
|
peptidylprolyl isomerase (cyclophilin)-like 6 |
| chr19_+_52800410 | 0.04 |
ENST00000595962.1
ENST00000598016.1 ENST00000334564.7 ENST00000490272.1 |
ZNF480
|
zinc finger protein 480 |
| chrX_-_153363125 | 0.04 |
ENST00000407218.1
ENST00000453960.2 |
MECP2
|
methyl CpG binding protein 2 (Rett syndrome) |
| chr14_-_99947121 | 0.04 |
ENST00000329331.3
ENST00000436070.2 |
SETD3
|
SET domain containing 3 |
| chr16_+_50776021 | 0.04 |
ENST00000566679.2
ENST00000564634.1 ENST00000398568.2 |
CYLD
|
cylindromatosis (turban tumor syndrome) |
| chr6_-_70506963 | 0.04 |
ENST00000370577.3
|
LMBRD1
|
LMBR1 domain containing 1 |
| chr5_+_118604439 | 0.04 |
ENST00000388882.5
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr1_-_43282945 | 0.04 |
ENST00000537227.1
|
CCDC23
|
coiled-coil domain containing 23 |
| chr12_+_77158021 | 0.04 |
ENST00000550876.1
|
ZDHHC17
|
zinc finger, DHHC-type containing 17 |
| chr2_-_39347524 | 0.04 |
ENST00000395038.2
ENST00000402219.2 |
SOS1
|
son of sevenless homolog 1 (Drosophila) |
| chr6_-_131384373 | 0.04 |
ENST00000392427.3
ENST00000525271.1 ENST00000527411.1 |
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr8_-_101322132 | 0.04 |
ENST00000523481.1
|
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr5_+_74807886 | 0.04 |
ENST00000514296.1
|
POLK
|
polymerase (DNA directed) kappa |
| chr2_-_136288740 | 0.04 |
ENST00000264159.6
ENST00000536680.1 |
ZRANB3
|
zinc finger, RAN-binding domain containing 3 |
| chr13_-_74708372 | 0.04 |
ENST00000377666.4
|
KLF12
|
Kruppel-like factor 12 |
| chr13_-_52026730 | 0.04 |
ENST00000420668.2
|
INTS6
|
integrator complex subunit 6 |
| chr10_-_112678976 | 0.04 |
ENST00000448814.1
|
BBIP1
|
BBSome interacting protein 1 |
| chr13_-_114018400 | 0.04 |
ENST00000375430.4
ENST00000375431.4 |
GRTP1
|
growth hormone regulated TBC protein 1 |
| chr10_+_76871229 | 0.04 |
ENST00000372690.3
|
SAMD8
|
sterile alpha motif domain containing 8 |
| chr7_-_33102399 | 0.04 |
ENST00000242210.7
|
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr15_+_90931450 | 0.04 |
ENST00000268182.5
ENST00000560738.1 ENST00000560418.1 |
IQGAP1
|
IQ motif containing GTPase activating protein 1 |
| chrX_+_14891522 | 0.04 |
ENST00000380492.3
ENST00000482354.1 |
MOSPD2
|
motile sperm domain containing 2 |
| chr12_+_50017327 | 0.04 |
ENST00000261897.1
|
PRPF40B
|
PRP40 pre-mRNA processing factor 40 homolog B (S. cerevisiae) |
| chr20_-_524362 | 0.04 |
ENST00000460062.2
ENST00000608066.1 |
CSNK2A1
|
casein kinase 2, alpha 1 polypeptide |
Network of associatons between targets according to the STRING database.
First level regulatory network of SP1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.0 | 0.1 | GO:0072573 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) negative regulation of CD40 signaling pathway(GO:2000349) |
| 0.0 | 0.0 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.0 | 0.1 | GO:1902232 | regulation of positive thymic T cell selection(GO:1902232) |
| 0.0 | 0.1 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.0 | 0.2 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.1 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.0 | 0.1 | GO:0097195 | pilomotor reflex(GO:0097195) |
| 0.0 | 0.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 | 0.1 | GO:0006679 | glucosylceramide biosynthetic process(GO:0006679) |
| 0.0 | 0.0 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.0 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.0 | GO:1903170 | negative regulation of calcium ion transmembrane transport(GO:1903170) |
| 0.0 | 0.0 | GO:0051450 | myoblast proliferation(GO:0051450) regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 0.1 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.0 | 0.0 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.0 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.0 | 0.0 | GO:0072171 | ureteric bud morphogenesis(GO:0060675) nephron tubule morphogenesis(GO:0072078) nephron epithelium morphogenesis(GO:0072088) mesonephric tubule morphogenesis(GO:0072171) |
| 0.0 | 0.0 | GO:0035750 | protein localization to myelin sheath abaxonal region(GO:0035750) |
| 0.0 | 0.1 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.0 | 0.0 | GO:1903515 | calcium ion transport from cytosol to endoplasmic reticulum(GO:1903515) |
| 0.0 | 0.1 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.0 | GO:2000417 | negative regulation of eosinophil migration(GO:2000417) |
| 0.0 | 0.1 | GO:0033504 | floor plate development(GO:0033504) |
| 0.0 | 0.1 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.0 | 0.1 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.0 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.0 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.1 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.0 | 0.1 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.0 | 0.0 | GO:0021769 | orbitofrontal cortex development(GO:0021769) |
| 0.0 | 0.1 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.0 | GO:0003064 | regulation of heart rate by hormone(GO:0003064) |
| 0.0 | 0.1 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 | 0.0 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.0 | 0.0 | GO:1903450 | regulation of G1 to G0 transition(GO:1903450) positive regulation of G1 to G0 transition(GO:1903452) |
| 0.0 | 0.1 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.1 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.0 | 0.0 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.0 | 0.0 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.0 | 0.1 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.0 | GO:0060178 | regulation of exocyst assembly(GO:0001928) regulation of exocyst localization(GO:0060178) |
| 0.0 | 0.0 | GO:0035822 | meiotic gene conversion(GO:0006311) gene conversion(GO:0035822) |
| 0.0 | 0.0 | GO:0003360 | brainstem development(GO:0003360) |
| 0.0 | 0.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.0 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 0.1 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.0 | 0.1 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.1 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.1 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.0 | GO:0014004 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.0 | 0.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.0 | GO:0035508 | positive regulation of myosin-light-chain-phosphatase activity(GO:0035508) |
| 0.0 | 0.0 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 0.0 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.1 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.0 | 0.0 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.0 | 0.0 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.0 | 0.0 | GO:0033488 | cholesterol biosynthetic process via 24,25-dihydrolanosterol(GO:0033488) |
| 0.0 | 0.0 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.0 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.0 | 0.1 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.0 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.0 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 0.0 | 0.0 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1903293 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 0.1 | GO:0075341 | host cell PML body(GO:0075341) |
| 0.0 | 0.1 | GO:0031213 | RSF complex(GO:0031213) |
| 0.0 | 0.0 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.0 | GO:0098796 | membrane protein complex(GO:0098796) |
| 0.0 | 0.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.0 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.1 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 0.1 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.1 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.0 | GO:0034676 | integrin alpha6-beta4 complex(GO:0034676) |
| 0.0 | 0.1 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.0 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.0 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0034678 | integrin alpha8-beta1 complex(GO:0034678) |
| 0.0 | 0.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.0 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 0.0 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.0 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) postsynaptic endocytic zone membrane(GO:0098844) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.1 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.0 | 0.1 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.1 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.0 | 0.1 | GO:0047322 | [hydroxymethylglutaryl-CoA reductase (NADPH)] kinase activity(GO:0047322) [acetyl-CoA carboxylase] kinase activity(GO:0050405) |
| 0.0 | 0.0 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.0 | 0.0 | GO:0086039 | lutropin-choriogonadotropic hormone receptor binding(GO:0031775) calcium-transporting ATPase activity involved in regulation of cardiac muscle cell membrane potential(GO:0086039) |
| 0.0 | 0.1 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.1 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.0 | 0.1 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.0 | 0.1 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.0 | 0.1 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.0 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.0 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.0 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.0 | GO:0000224 | peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase activity(GO:0000224) |
| 0.0 | 0.0 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.0 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.0 | 0.1 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.0 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.0 | 0.0 | GO:0000406 | double-strand/single-strand DNA junction binding(GO:0000406) dinucleotide repeat insertion binding(GO:0032181) |
| 0.0 | 0.0 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.0 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.1 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.0 | 0.0 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.0 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 0.0 | GO:0016768 | spermine synthase activity(GO:0016768) |
| 0.0 | 0.0 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.0 | 0.0 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.0 | GO:0008398 | sterol 14-demethylase activity(GO:0008398) |
| 0.0 | 0.0 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.0 | GO:0050664 | oxidoreductase activity, acting on NAD(P)H, oxygen as acceptor(GO:0050664) |
| 0.0 | 0.0 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 0.1 | REACTOME RNA POL I TRANSCRIPTION | Genes involved in RNA Polymerase I Transcription |
| 0.0 | 0.0 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |