Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
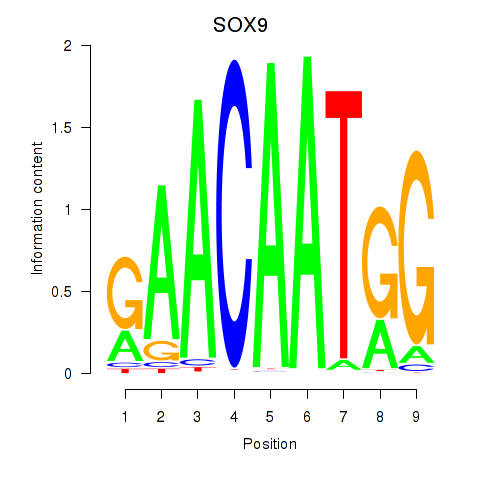
Results for SOX9
Z-value: 0.51
Transcription factors associated with SOX9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX9
|
ENSG00000125398.5 | SRY-box transcription factor 9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX9 | hg19_v2_chr17_+_70117153_70117174 | 0.88 | 1.2e-01 | Click! |
Activity profile of SOX9 motif
Sorted Z-values of SOX9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_77167376 | 0.22 |
ENST00000435495.2
|
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr12_-_120805872 | 0.21 |
ENST00000546985.1
|
MSI1
|
musashi RNA-binding protein 1 |
| chr14_+_52456327 | 0.20 |
ENST00000556760.1
|
C14orf166
|
chromosome 14 open reading frame 166 |
| chr7_+_77167343 | 0.19 |
ENST00000433369.2
ENST00000415482.2 |
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr1_-_26232522 | 0.16 |
ENST00000399728.1
|
STMN1
|
stathmin 1 |
| chr11_-_46848393 | 0.16 |
ENST00000526496.1
|
CKAP5
|
cytoskeleton associated protein 5 |
| chr17_-_73149921 | 0.16 |
ENST00000481647.1
ENST00000470924.1 |
HN1
|
hematological and neurological expressed 1 |
| chr14_-_51027838 | 0.16 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr8_+_79578282 | 0.16 |
ENST00000263849.4
|
ZC2HC1A
|
zinc finger, C2HC-type containing 1A |
| chr17_-_37764128 | 0.15 |
ENST00000302584.4
|
NEUROD2
|
neuronal differentiation 2 |
| chr22_-_42739533 | 0.14 |
ENST00000515426.1
|
TCF20
|
transcription factor 20 (AR1) |
| chr9_-_35828576 | 0.14 |
ENST00000377984.2
ENST00000423537.2 ENST00000472182.1 |
FAM221B
|
family with sequence similarity 221, member B |
| chr2_-_55277692 | 0.14 |
ENST00000394611.2
|
RTN4
|
reticulon 4 |
| chr3_-_185826718 | 0.13 |
ENST00000413301.1
ENST00000421809.1 |
ETV5
|
ets variant 5 |
| chr6_-_108278456 | 0.13 |
ENST00000429168.1
|
SEC63
|
SEC63 homolog (S. cerevisiae) |
| chr1_+_26503894 | 0.13 |
ENST00000361530.6
ENST00000374253.5 |
CNKSR1
|
connector enhancer of kinase suppressor of Ras 1 |
| chr14_+_93357749 | 0.13 |
ENST00000557689.1
|
RP11-862G15.1
|
RP11-862G15.1 |
| chr1_-_26232951 | 0.13 |
ENST00000426559.2
ENST00000455785.2 |
STMN1
|
stathmin 1 |
| chr16_-_71918033 | 0.13 |
ENST00000425432.1
ENST00000313565.6 ENST00000568666.1 ENST00000562797.1 ENST00000564134.1 |
ZNF821
|
zinc finger protein 821 |
| chr17_+_27573875 | 0.13 |
ENST00000225387.3
|
CRYBA1
|
crystallin, beta A1 |
| chr3_-_71777824 | 0.13 |
ENST00000469524.1
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr3_+_187871659 | 0.12 |
ENST00000416784.1
ENST00000430340.1 ENST00000414139.1 ENST00000454789.1 |
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr2_-_55276320 | 0.12 |
ENST00000357376.3
|
RTN4
|
reticulon 4 |
| chr12_+_113495492 | 0.12 |
ENST00000257600.3
|
DTX1
|
deltex homolog 1 (Drosophila) |
| chr21_-_19191703 | 0.12 |
ENST00000284881.4
ENST00000400559.3 ENST00000400558.3 |
C21orf91
|
chromosome 21 open reading frame 91 |
| chr11_-_95522907 | 0.12 |
ENST00000358780.5
ENST00000542135.1 |
FAM76B
|
family with sequence similarity 76, member B |
| chr7_-_27205136 | 0.12 |
ENST00000396345.1
ENST00000343483.6 |
HOXA9
|
homeobox A9 |
| chr11_-_76155618 | 0.12 |
ENST00000530759.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr5_-_159546396 | 0.11 |
ENST00000523662.1
ENST00000456329.3 ENST00000307063.7 |
PWWP2A
|
PWWP domain containing 2A |
| chr10_+_91461337 | 0.11 |
ENST00000260753.4
ENST00000416354.1 ENST00000394289.2 ENST00000371728.3 |
KIF20B
|
kinesin family member 20B |
| chr12_-_122985067 | 0.11 |
ENST00000540586.1
ENST00000543897.1 |
ZCCHC8
|
zinc finger, CCHC domain containing 8 |
| chr1_+_185014496 | 0.11 |
ENST00000367510.3
|
RNF2
|
ring finger protein 2 |
| chr2_+_170590321 | 0.11 |
ENST00000392647.2
|
KLHL23
|
kelch-like family member 23 |
| chr19_+_8455200 | 0.11 |
ENST00000601897.1
ENST00000594216.1 |
RAB11B
|
RAB11B, member RAS oncogene family |
| chr5_-_65018834 | 0.11 |
ENST00000506816.1
|
SGTB
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr11_-_95523500 | 0.11 |
ENST00000540054.1
|
FAM76B
|
family with sequence similarity 76, member B |
| chrX_-_20159934 | 0.10 |
ENST00000379593.1
ENST00000379607.5 |
EIF1AX
|
eukaryotic translation initiation factor 1A, X-linked |
| chr2_-_55277436 | 0.10 |
ENST00000354474.6
|
RTN4
|
reticulon 4 |
| chr3_-_57583052 | 0.10 |
ENST00000496292.1
ENST00000489843.1 |
ARF4
|
ADP-ribosylation factor 4 |
| chr8_-_80993010 | 0.10 |
ENST00000537855.1
ENST00000520527.1 ENST00000517427.1 ENST00000448733.2 ENST00000379097.3 |
TPD52
|
tumor protein D52 |
| chr17_+_30813576 | 0.10 |
ENST00000313401.3
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
| chr10_+_91461413 | 0.10 |
ENST00000447580.1
|
KIF20B
|
kinesin family member 20B |
| chr1_-_243418344 | 0.10 |
ENST00000366542.1
|
CEP170
|
centrosomal protein 170kDa |
| chr14_+_52456193 | 0.10 |
ENST00000261700.3
|
C14orf166
|
chromosome 14 open reading frame 166 |
| chr13_+_53029564 | 0.10 |
ENST00000468284.1
ENST00000378034.3 ENST00000258607.5 ENST00000378037.5 |
CKAP2
|
cytoskeleton associated protein 2 |
| chr1_-_26233423 | 0.10 |
ENST00000357865.2
|
STMN1
|
stathmin 1 |
| chr6_-_27440460 | 0.10 |
ENST00000377419.1
|
ZNF184
|
zinc finger protein 184 |
| chr8_+_98656693 | 0.09 |
ENST00000519934.1
|
MTDH
|
metadherin |
| chr5_-_43313574 | 0.09 |
ENST00000325110.6
ENST00000433297.2 |
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr14_+_32546274 | 0.09 |
ENST00000396582.2
|
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr20_-_17662705 | 0.09 |
ENST00000455029.2
|
RRBP1
|
ribosome binding protein 1 |
| chr7_-_112635675 | 0.09 |
ENST00000447785.1
ENST00000451962.1 |
AC018464.3
|
AC018464.3 |
| chr6_-_27440837 | 0.09 |
ENST00000211936.6
|
ZNF184
|
zinc finger protein 184 |
| chr1_-_243418621 | 0.09 |
ENST00000366544.1
ENST00000366543.1 |
CEP170
|
centrosomal protein 170kDa |
| chr4_+_113152978 | 0.09 |
ENST00000309703.6
|
AP1AR
|
adaptor-related protein complex 1 associated regulatory protein |
| chr7_-_27239703 | 0.09 |
ENST00000222753.4
|
HOXA13
|
homeobox A13 |
| chr2_+_46926326 | 0.09 |
ENST00000394861.2
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr19_+_58790314 | 0.09 |
ENST00000196548.5
ENST00000608843.1 |
ZNF8
ZNF8
|
Zinc finger protein 8 zinc finger protein 8 |
| chr6_-_86353510 | 0.09 |
ENST00000444272.1
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chrX_+_135579238 | 0.09 |
ENST00000535601.1
ENST00000448450.1 ENST00000425695.1 |
HTATSF1
|
HIV-1 Tat specific factor 1 |
| chr1_+_200993071 | 0.09 |
ENST00000446333.1
ENST00000458003.1 |
RP11-168O16.1
|
RP11-168O16.1 |
| chr5_+_74632993 | 0.09 |
ENST00000287936.4
|
HMGCR
|
3-hydroxy-3-methylglutaryl-CoA reductase |
| chr16_-_15474904 | 0.09 |
ENST00000534094.1
|
NPIPA5
|
nuclear pore complex interacting protein family, member A5 |
| chr4_+_24661479 | 0.09 |
ENST00000569621.1
|
RP11-496D24.2
|
RP11-496D24.2 |
| chr1_+_25071848 | 0.09 |
ENST00000374379.4
|
CLIC4
|
chloride intracellular channel 4 |
| chr12_+_95611516 | 0.09 |
ENST00000436874.1
|
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr2_-_70475701 | 0.09 |
ENST00000282574.4
|
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr15_+_52311398 | 0.09 |
ENST00000261845.5
|
MAPK6
|
mitogen-activated protein kinase 6 |
| chr3_-_149051444 | 0.09 |
ENST00000296059.2
|
TM4SF18
|
transmembrane 4 L six family member 18 |
| chr1_-_40367668 | 0.09 |
ENST00000397332.2
ENST00000429311.1 |
MYCL
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr2_+_61293021 | 0.09 |
ENST00000402291.1
|
KIAA1841
|
KIAA1841 |
| chr11_-_102323740 | 0.09 |
ENST00000398136.2
|
TMEM123
|
transmembrane protein 123 |
| chr4_+_113152881 | 0.08 |
ENST00000274000.5
|
AP1AR
|
adaptor-related protein complex 1 associated regulatory protein |
| chr4_+_54243917 | 0.08 |
ENST00000507166.1
|
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr15_+_93426514 | 0.08 |
ENST00000556722.1
|
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr17_-_56605341 | 0.08 |
ENST00000583114.1
|
SEPT4
|
septin 4 |
| chr12_-_91546926 | 0.08 |
ENST00000550758.1
|
DCN
|
decorin |
| chr12_-_95611149 | 0.08 |
ENST00000549499.1
ENST00000343958.4 ENST00000546711.1 |
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr6_+_37787704 | 0.08 |
ENST00000474522.1
|
ZFAND3
|
zinc finger, AN1-type domain 3 |
| chr1_-_53018654 | 0.08 |
ENST00000257177.4
ENST00000355809.4 ENST00000528642.1 ENST00000470626.1 ENST00000371544.3 |
ZCCHC11
|
zinc finger, CCHC domain containing 11 |
| chrX_-_15683147 | 0.08 |
ENST00000380342.3
|
TMEM27
|
transmembrane protein 27 |
| chr6_+_56911476 | 0.08 |
ENST00000545356.1
|
KIAA1586
|
KIAA1586 |
| chr5_+_74633036 | 0.08 |
ENST00000343975.5
|
HMGCR
|
3-hydroxy-3-methylglutaryl-CoA reductase |
| chr2_-_37899323 | 0.08 |
ENST00000295324.3
ENST00000457889.1 |
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr11_+_125703204 | 0.08 |
ENST00000534411.1
ENST00000457514.2 |
PATE4
|
prostate and testis expressed 4 |
| chr14_-_65569057 | 0.08 |
ENST00000555419.1
ENST00000341653.2 |
MAX
|
MYC associated factor X |
| chr15_+_41624892 | 0.08 |
ENST00000260359.6
ENST00000450318.1 ENST00000450592.2 ENST00000559596.1 ENST00000414849.2 ENST00000560747.1 ENST00000560177.1 |
NUSAP1
|
nucleolar and spindle associated protein 1 |
| chr12_+_69979446 | 0.08 |
ENST00000543146.2
|
CCT2
|
chaperonin containing TCP1, subunit 2 (beta) |
| chr1_-_93645818 | 0.08 |
ENST00000370280.1
ENST00000479918.1 |
TMED5
|
transmembrane emp24 protein transport domain containing 5 |
| chr11_+_63953587 | 0.08 |
ENST00000305218.4
ENST00000538945.1 |
STIP1
|
stress-induced-phosphoprotein 1 |
| chr3_-_88108192 | 0.08 |
ENST00000309534.6
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr14_-_91884115 | 0.08 |
ENST00000389857.6
|
CCDC88C
|
coiled-coil domain containing 88C |
| chr12_+_56435637 | 0.08 |
ENST00000356464.5
ENST00000552361.1 |
RPS26
|
ribosomal protein S26 |
| chr5_-_39425222 | 0.08 |
ENST00000320816.6
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr19_+_45971246 | 0.08 |
ENST00000585836.1
ENST00000417353.2 ENST00000353609.3 ENST00000591858.1 ENST00000443841.2 ENST00000590335.1 |
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B |
| chr2_+_54683419 | 0.08 |
ENST00000356805.4
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1 |
| chr18_+_20494078 | 0.08 |
ENST00000579124.1
ENST00000577588.1 ENST00000582354.1 ENST00000581819.1 |
RBBP8
|
retinoblastoma binding protein 8 |
| chr12_-_122985494 | 0.08 |
ENST00000336229.4
|
ZCCHC8
|
zinc finger, CCHC domain containing 8 |
| chr11_-_102323489 | 0.08 |
ENST00000361236.3
|
TMEM123
|
transmembrane protein 123 |
| chr9_-_127905736 | 0.07 |
ENST00000336505.6
ENST00000373549.4 |
SCAI
|
suppressor of cancer cell invasion |
| chr2_-_201828356 | 0.07 |
ENST00000234296.2
|
ORC2
|
origin recognition complex, subunit 2 |
| chr17_-_57232525 | 0.07 |
ENST00000583380.1
ENST00000580541.1 ENST00000578105.1 ENST00000437036.2 |
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr10_+_6244829 | 0.07 |
ENST00000317350.4
ENST00000379785.1 ENST00000379782.3 ENST00000360521.2 ENST00000379775.4 |
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr22_+_38864041 | 0.07 |
ENST00000216014.4
ENST00000409006.3 |
KDELR3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chr7_+_155090271 | 0.07 |
ENST00000476756.1
|
INSIG1
|
insulin induced gene 1 |
| chr12_+_95611569 | 0.07 |
ENST00000261219.6
ENST00000551472.1 ENST00000552821.1 |
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr15_-_52263937 | 0.07 |
ENST00000315141.5
ENST00000299601.5 |
LEO1
|
Leo1, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae) |
| chr9_+_103235365 | 0.07 |
ENST00000374879.4
|
TMEFF1
|
transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr5_-_168006324 | 0.07 |
ENST00000522176.1
|
PANK3
|
pantothenate kinase 3 |
| chrX_-_15872914 | 0.07 |
ENST00000380291.1
ENST00000545766.1 ENST00000421527.2 ENST00000329235.2 |
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr8_-_38325219 | 0.07 |
ENST00000533668.1
ENST00000413133.2 ENST00000397108.4 ENST00000526742.1 ENST00000525001.1 ENST00000425967.3 ENST00000529552.1 ENST00000397113.2 |
FGFR1
|
fibroblast growth factor receptor 1 |
| chr19_+_58514229 | 0.07 |
ENST00000546949.1
ENST00000553254.1 ENST00000547364.1 |
CTD-2368P22.1
|
HCG1811579; Uncharacterized protein |
| chr1_-_40367530 | 0.07 |
ENST00000372816.2
ENST00000372815.1 |
MYCL
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr11_-_46867780 | 0.07 |
ENST00000529230.1
ENST00000415402.1 ENST00000312055.5 |
CKAP5
|
cytoskeleton associated protein 5 |
| chr12_+_69979113 | 0.07 |
ENST00000299300.6
|
CCT2
|
chaperonin containing TCP1, subunit 2 (beta) |
| chr11_+_117103441 | 0.07 |
ENST00000531287.1
ENST00000531452.1 |
RNF214
|
ring finger protein 214 |
| chr11_+_34073195 | 0.07 |
ENST00000341394.4
|
CAPRIN1
|
cell cycle associated protein 1 |
| chr10_-_94050820 | 0.07 |
ENST00000265997.4
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr6_-_24877490 | 0.07 |
ENST00000540914.1
ENST00000378023.4 |
FAM65B
|
family with sequence similarity 65, member B |
| chr20_-_40247002 | 0.07 |
ENST00000373222.3
|
CHD6
|
chromodomain helicase DNA binding protein 6 |
| chr2_-_175869936 | 0.07 |
ENST00000409900.3
|
CHN1
|
chimerin 1 |
| chr1_+_209602156 | 0.07 |
ENST00000429156.1
ENST00000366437.3 ENST00000603283.1 ENST00000431096.1 |
MIR205HG
|
MIR205 host gene (non-protein coding) |
| chr11_-_77531858 | 0.07 |
ENST00000360355.2
|
RSF1
|
remodeling and spacing factor 1 |
| chr12_-_45270151 | 0.07 |
ENST00000429094.2
|
NELL2
|
NEL-like 2 (chicken) |
| chr4_+_95128748 | 0.07 |
ENST00000359052.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr11_-_69519410 | 0.07 |
ENST00000294312.3
|
FGF19
|
fibroblast growth factor 19 |
| chr15_-_56035177 | 0.07 |
ENST00000389286.4
ENST00000561292.1 |
PRTG
|
protogenin |
| chr6_+_131571535 | 0.07 |
ENST00000474850.2
|
AKAP7
|
A kinase (PRKA) anchor protein 7 |
| chr3_+_44596679 | 0.07 |
ENST00000426540.1
ENST00000431636.1 ENST00000341840.3 ENST00000273320.3 |
ZKSCAN7
|
zinc finger with KRAB and SCAN domains 7 |
| chr2_+_46926048 | 0.07 |
ENST00000306503.5
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chrX_-_130037162 | 0.07 |
ENST00000432489.1
|
ENOX2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr1_-_182360498 | 0.07 |
ENST00000417584.2
|
GLUL
|
glutamate-ammonia ligase |
| chr11_-_95522639 | 0.07 |
ENST00000536839.1
|
FAM76B
|
family with sequence similarity 76, member B |
| chr1_-_26231589 | 0.07 |
ENST00000374291.1
|
STMN1
|
stathmin 1 |
| chr7_+_16793160 | 0.07 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13 |
| chr3_-_149688502 | 0.07 |
ENST00000481767.1
ENST00000475518.1 |
PFN2
|
profilin 2 |
| chr1_-_154164534 | 0.07 |
ENST00000271850.7
ENST00000368530.2 |
TPM3
|
tropomyosin 3 |
| chr14_+_102276209 | 0.07 |
ENST00000445439.3
ENST00000334743.5 ENST00000557095.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr14_-_103989033 | 0.07 |
ENST00000553878.1
ENST00000557530.1 |
CKB
|
creatine kinase, brain |
| chr14_+_63671105 | 0.07 |
ENST00000316754.3
|
RHOJ
|
ras homolog family member J |
| chr15_-_59041768 | 0.07 |
ENST00000402627.1
ENST00000396140.2 ENST00000559053.1 ENST00000561288.1 |
ADAM10
|
ADAM metallopeptidase domain 10 |
| chr4_+_95129061 | 0.06 |
ENST00000354268.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr1_+_64239657 | 0.06 |
ENST00000371080.1
ENST00000371079.1 |
ROR1
|
receptor tyrosine kinase-like orphan receptor 1 |
| chr11_+_34073269 | 0.06 |
ENST00000389645.3
|
CAPRIN1
|
cell cycle associated protein 1 |
| chr4_+_54243862 | 0.06 |
ENST00000306932.6
|
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr8_-_101965559 | 0.06 |
ENST00000353245.3
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr9_+_108463234 | 0.06 |
ENST00000374688.1
|
TMEM38B
|
transmembrane protein 38B |
| chr5_+_148521454 | 0.06 |
ENST00000508983.1
|
ABLIM3
|
actin binding LIM protein family, member 3 |
| chr17_+_26646121 | 0.06 |
ENST00000226230.6
|
TMEM97
|
transmembrane protein 97 |
| chr8_+_133787586 | 0.06 |
ENST00000395379.1
ENST00000395386.2 ENST00000337920.4 |
PHF20L1
|
PHD finger protein 20-like 1 |
| chr20_-_40247133 | 0.06 |
ENST00000373233.3
ENST00000309279.7 |
CHD6
|
chromodomain helicase DNA binding protein 6 |
| chr3_+_51422478 | 0.06 |
ENST00000528157.1
|
MANF
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr6_+_64282447 | 0.06 |
ENST00000370650.2
ENST00000578299.1 |
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr7_+_120590803 | 0.06 |
ENST00000315870.5
ENST00000339121.5 ENST00000445699.1 |
ING3
|
inhibitor of growth family, member 3 |
| chr18_+_7946941 | 0.06 |
ENST00000444013.1
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M |
| chr4_-_76598326 | 0.06 |
ENST00000503660.1
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr19_-_8070474 | 0.06 |
ENST00000407627.2
ENST00000593807.1 |
ELAVL1
|
ELAV like RNA binding protein 1 |
| chr2_+_109271481 | 0.06 |
ENST00000542845.1
ENST00000393314.2 |
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr3_-_149051194 | 0.06 |
ENST00000470080.1
|
TM4SF18
|
transmembrane 4 L six family member 18 |
| chr7_+_120591170 | 0.06 |
ENST00000431467.1
|
ING3
|
inhibitor of growth family, member 3 |
| chr17_+_16945820 | 0.06 |
ENST00000577514.1
|
MPRIP
|
myosin phosphatase Rho interacting protein |
| chr14_+_53019993 | 0.06 |
ENST00000542169.2
ENST00000555622.1 |
GPR137C
|
G protein-coupled receptor 137C |
| chr14_+_59655369 | 0.06 |
ENST00000360909.3
ENST00000351081.1 ENST00000556135.1 |
DAAM1
|
dishevelled associated activator of morphogenesis 1 |
| chrX_-_45060135 | 0.06 |
ENST00000398000.2
ENST00000377934.4 |
CXorf36
|
chromosome X open reading frame 36 |
| chr1_-_113247543 | 0.06 |
ENST00000414971.1
ENST00000534717.1 |
RHOC
|
ras homolog family member C |
| chrX_-_130037198 | 0.06 |
ENST00000370935.1
ENST00000338144.3 ENST00000394363.1 |
ENOX2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr20_+_42086525 | 0.06 |
ENST00000244020.3
|
SRSF6
|
serine/arginine-rich splicing factor 6 |
| chr9_-_85882145 | 0.06 |
ENST00000328788.1
|
FRMD3
|
FERM domain containing 3 |
| chr5_+_179135246 | 0.06 |
ENST00000508787.1
|
CANX
|
calnexin |
| chr11_+_117103333 | 0.06 |
ENST00000534428.1
|
RNF214
|
ring finger protein 214 |
| chr2_-_61765315 | 0.06 |
ENST00000406957.1
ENST00000401558.2 |
XPO1
|
exportin 1 (CRM1 homolog, yeast) |
| chr4_-_83351005 | 0.06 |
ENST00000295470.5
|
HNRNPDL
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr17_+_75447326 | 0.06 |
ENST00000591088.1
|
SEPT9
|
septin 9 |
| chr17_+_74723031 | 0.06 |
ENST00000586200.1
|
METTL23
|
methyltransferase like 23 |
| chr3_+_43328004 | 0.06 |
ENST00000454177.1
ENST00000429705.2 ENST00000296088.7 ENST00000437827.1 |
SNRK
|
SNF related kinase |
| chr10_+_49514698 | 0.06 |
ENST00000432379.1
ENST00000429041.1 ENST00000374189.1 |
MAPK8
|
mitogen-activated protein kinase 8 |
| chr5_+_138678131 | 0.06 |
ENST00000394795.2
ENST00000510080.1 |
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr14_+_32546145 | 0.06 |
ENST00000556611.1
ENST00000539826.2 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr7_+_134464414 | 0.06 |
ENST00000361901.2
|
CALD1
|
caldesmon 1 |
| chr19_+_54641444 | 0.06 |
ENST00000221232.5
ENST00000358389.3 |
CNOT3
|
CCR4-NOT transcription complex, subunit 3 |
| chr15_+_71184931 | 0.06 |
ENST00000560369.1
ENST00000260382.5 |
LRRC49
|
leucine rich repeat containing 49 |
| chr7_+_128379449 | 0.06 |
ENST00000479257.1
|
CALU
|
calumenin |
| chr7_-_148725733 | 0.06 |
ENST00000286091.4
|
PDIA4
|
protein disulfide isomerase family A, member 4 |
| chr3_-_182698381 | 0.06 |
ENST00000292782.4
|
DCUN1D1
|
DCN1, defective in cullin neddylation 1, domain containing 1 |
| chr10_-_128359074 | 0.06 |
ENST00000544758.1
|
C10orf90
|
chromosome 10 open reading frame 90 |
| chr6_-_11232891 | 0.06 |
ENST00000379433.5
ENST00000379446.5 |
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr4_+_71570430 | 0.06 |
ENST00000417478.2
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr7_+_76751926 | 0.06 |
ENST00000285871.4
ENST00000431197.1 |
CCDC146
|
coiled-coil domain containing 146 |
| chr19_+_8455077 | 0.06 |
ENST00000328024.6
|
RAB11B
|
RAB11B, member RAS oncogene family |
| chr12_+_69979210 | 0.06 |
ENST00000544368.2
|
CCT2
|
chaperonin containing TCP1, subunit 2 (beta) |
| chr14_-_91976488 | 0.06 |
ENST00000554684.1
ENST00000337238.4 ENST00000428424.2 ENST00000554511.1 |
SMEK1
|
SMEK homolog 1, suppressor of mek1 (Dictyostelium) |
| chr4_-_83351294 | 0.06 |
ENST00000502762.1
|
HNRNPDL
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr1_+_14075865 | 0.06 |
ENST00000413440.1
|
PRDM2
|
PR domain containing 2, with ZNF domain |
| chr11_+_13299186 | 0.06 |
ENST00000527998.1
ENST00000396441.3 ENST00000533520.1 ENST00000529825.1 ENST00000389707.4 ENST00000401424.1 ENST00000529388.1 ENST00000530357.1 ENST00000403290.1 ENST00000361003.4 ENST00000389708.3 ENST00000403510.3 ENST00000482049.1 |
ARNTL
|
aryl hydrocarbon receptor nuclear translocator-like |
| chr12_-_45270077 | 0.06 |
ENST00000551601.1
ENST00000549027.1 ENST00000452445.2 |
NELL2
|
NEL-like 2 (chicken) |
| chr15_+_41136369 | 0.06 |
ENST00000563656.1
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr5_+_94890778 | 0.06 |
ENST00000380009.4
|
ARSK
|
arylsulfatase family, member K |
| chr6_-_27114577 | 0.06 |
ENST00000356950.1
ENST00000396891.4 |
HIST1H2BK
|
histone cluster 1, H2bk |
| chr8_-_101965146 | 0.06 |
ENST00000395957.2
ENST00000395948.2 ENST00000457309.1 |
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr3_-_16306432 | 0.05 |
ENST00000383775.4
ENST00000488423.1 |
DPH3
|
diphthamide biosynthesis 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX9
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.1 | 0.2 | GO:0051086 | chaperone mediated protein folding independent of cofactor(GO:0051086) |
| 0.1 | 0.2 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.0 | 0.1 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.4 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.2 | GO:1903436 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.0 | 0.1 | GO:0021718 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.0 | 0.2 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.1 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.1 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.1 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 | 0.2 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.0 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.0 | 0.1 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.0 | 0.4 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 0.1 | GO:0001835 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.2 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.1 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.0 | 0.1 | GO:2000583 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.0 | 0.1 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.0 | 0.1 | GO:1904617 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.0 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.0 | 0.2 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.3 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.2 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.1 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.0 | GO:2000417 | negative regulation of eosinophil migration(GO:2000417) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.2 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.0 | GO:2000586 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) |
| 0.0 | 0.1 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.1 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.0 | 0.0 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.0 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.2 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.1 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.0 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.0 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031213 | RSF complex(GO:0031213) |
| 0.0 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.1 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 0.1 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.0 | 0.4 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.1 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 0.1 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.0 | 0.1 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.0 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.0 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.0 | 0.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.0 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |