Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
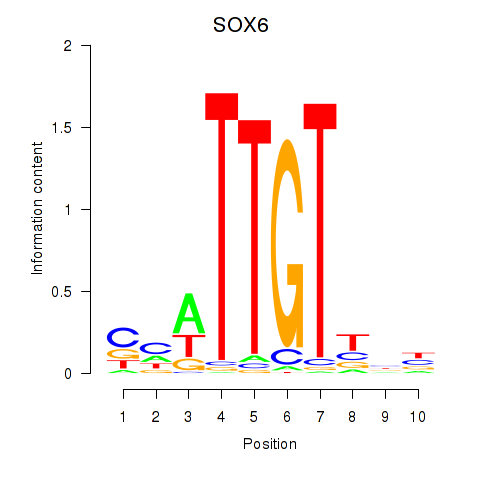
Results for SOX6
Z-value: 0.26
Transcription factors associated with SOX6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX6
|
ENSG00000110693.11 | SRY-box transcription factor 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX6 | hg19_v2_chr11_-_16430399_16430440 | 0.92 | 7.5e-02 | Click! |
Activity profile of SOX6 motif
Sorted Z-values of SOX6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_51027838 | 0.18 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr2_-_37899323 | 0.13 |
ENST00000295324.3
ENST00000457889.1 |
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr5_-_65018834 | 0.12 |
ENST00000506816.1
|
SGTB
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr15_+_101402041 | 0.12 |
ENST00000558475.1
ENST00000558641.1 ENST00000559673.1 |
RP11-66B24.1
|
RP11-66B24.1 |
| chr11_-_8892464 | 0.09 |
ENST00000527347.1
ENST00000526241.1 ENST00000526126.1 ENST00000530938.1 ENST00000526057.1 |
ST5
|
suppression of tumorigenicity 5 |
| chr1_-_26232951 | 0.08 |
ENST00000426559.2
ENST00000455785.2 |
STMN1
|
stathmin 1 |
| chr11_-_85430088 | 0.08 |
ENST00000533057.1
ENST00000533892.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr5_-_114515734 | 0.08 |
ENST00000514154.1
ENST00000282369.3 |
TRIM36
|
tripartite motif containing 36 |
| chr12_-_95611149 | 0.08 |
ENST00000549499.1
ENST00000343958.4 ENST00000546711.1 |
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr14_+_23791159 | 0.08 |
ENST00000557702.1
|
PABPN1
|
poly(A) binding protein, nuclear 1 |
| chr9_-_88896977 | 0.08 |
ENST00000311534.6
|
ISCA1
|
iron-sulfur cluster assembly 1 |
| chr3_-_167813132 | 0.08 |
ENST00000309027.4
|
GOLIM4
|
golgi integral membrane protein 4 |
| chr6_+_122720681 | 0.08 |
ENST00000368455.4
ENST00000452194.1 |
HSF2
|
heat shock transcription factor 2 |
| chr1_-_26233423 | 0.08 |
ENST00000357865.2
|
STMN1
|
stathmin 1 |
| chr14_-_91884115 | 0.08 |
ENST00000389857.6
|
CCDC88C
|
coiled-coil domain containing 88C |
| chr2_+_170590321 | 0.08 |
ENST00000392647.2
|
KLHL23
|
kelch-like family member 23 |
| chr6_-_108278456 | 0.07 |
ENST00000429168.1
|
SEC63
|
SEC63 homolog (S. cerevisiae) |
| chr7_+_99006232 | 0.07 |
ENST00000403633.2
|
BUD31
|
BUD31 homolog (S. cerevisiae) |
| chr2_-_42180940 | 0.07 |
ENST00000378711.2
|
C2orf91
|
chromosome 2 open reading frame 91 |
| chr11_-_85430204 | 0.07 |
ENST00000389958.3
ENST00000527794.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr1_-_154155675 | 0.07 |
ENST00000330188.9
ENST00000341485.5 |
TPM3
|
tropomyosin 3 |
| chr17_-_79849438 | 0.07 |
ENST00000331204.4
ENST00000505490.2 |
ALYREF
|
Aly/REF export factor |
| chr12_+_95611536 | 0.07 |
ENST00000549002.1
|
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr1_+_68150744 | 0.07 |
ENST00000370986.4
ENST00000370985.3 |
GADD45A
|
growth arrest and DNA-damage-inducible, alpha |
| chr17_-_56065484 | 0.07 |
ENST00000581208.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr1_-_26232522 | 0.06 |
ENST00000399728.1
|
STMN1
|
stathmin 1 |
| chr6_+_32937083 | 0.06 |
ENST00000456339.1
|
BRD2
|
bromodomain containing 2 |
| chr11_+_63974135 | 0.06 |
ENST00000544997.1
ENST00000345728.5 ENST00000279227.5 |
FERMT3
|
fermitin family member 3 |
| chr15_+_79166065 | 0.06 |
ENST00000559690.1
ENST00000559158.1 |
MORF4L1
|
mortality factor 4 like 1 |
| chr15_+_63335899 | 0.06 |
ENST00000561266.1
|
TPM1
|
tropomyosin 1 (alpha) |
| chr14_-_71107921 | 0.06 |
ENST00000553982.1
ENST00000500016.1 |
CTD-2540L5.5
CTD-2540L5.6
|
CTD-2540L5.5 CTD-2540L5.6 |
| chr11_-_102323740 | 0.06 |
ENST00000398136.2
|
TMEM123
|
transmembrane protein 123 |
| chr15_+_52311398 | 0.06 |
ENST00000261845.5
|
MAPK6
|
mitogen-activated protein kinase 6 |
| chr17_+_76037081 | 0.06 |
ENST00000588549.1
|
TNRC6C
|
trinucleotide repeat containing 6C |
| chr10_+_35416223 | 0.06 |
ENST00000489321.1
ENST00000427847.2 ENST00000345491.3 ENST00000395895.2 ENST00000374728.3 ENST00000487132.1 |
CREM
|
cAMP responsive element modulator |
| chr2_-_160472052 | 0.06 |
ENST00000437839.1
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr1_+_14075865 | 0.06 |
ENST00000413440.1
|
PRDM2
|
PR domain containing 2, with ZNF domain |
| chr15_+_52043758 | 0.06 |
ENST00000249700.4
ENST00000539962.2 |
TMOD2
|
tropomodulin 2 (neuronal) |
| chr12_+_95611569 | 0.06 |
ENST00000261219.6
ENST00000551472.1 ENST00000552821.1 |
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr12_+_95611516 | 0.06 |
ENST00000436874.1
|
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr4_-_146101304 | 0.06 |
ENST00000447906.2
|
OTUD4
|
OTU domain containing 4 |
| chr5_-_39425222 | 0.05 |
ENST00000320816.6
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr4_+_54243917 | 0.05 |
ENST00000507166.1
|
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr17_+_52978156 | 0.05 |
ENST00000348161.4
|
TOM1L1
|
target of myb1 (chicken)-like 1 |
| chr5_-_180018540 | 0.05 |
ENST00000292641.3
|
SCGB3A1
|
secretoglobin, family 3A, member 1 |
| chr5_-_16936340 | 0.05 |
ENST00000507288.1
ENST00000513610.1 |
MYO10
|
myosin X |
| chr2_+_61293021 | 0.05 |
ENST00000402291.1
|
KIAA1841
|
KIAA1841 |
| chr11_-_102323489 | 0.05 |
ENST00000361236.3
|
TMEM123
|
transmembrane protein 123 |
| chr1_+_150229554 | 0.05 |
ENST00000369111.4
|
CA14
|
carbonic anhydrase XIV |
| chr1_+_228337553 | 0.05 |
ENST00000366714.2
|
GJC2
|
gap junction protein, gamma 2, 47kDa |
| chr2_+_153191706 | 0.05 |
ENST00000288670.9
|
FMNL2
|
formin-like 2 |
| chr1_+_47799542 | 0.05 |
ENST00000471289.2
ENST00000450808.2 |
CMPK1
|
cytidine monophosphate (UMP-CMP) kinase 1, cytosolic |
| chr5_-_76788134 | 0.05 |
ENST00000507029.1
|
WDR41
|
WD repeat domain 41 |
| chr12_+_69979446 | 0.05 |
ENST00000543146.2
|
CCT2
|
chaperonin containing TCP1, subunit 2 (beta) |
| chr2_-_55276320 | 0.05 |
ENST00000357376.3
|
RTN4
|
reticulon 4 |
| chr5_+_110073853 | 0.05 |
ENST00000513807.1
ENST00000509442.2 |
SLC25A46
|
solute carrier family 25, member 46 |
| chr2_+_46926326 | 0.05 |
ENST00000394861.2
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr18_+_20494078 | 0.05 |
ENST00000579124.1
ENST00000577588.1 ENST00000582354.1 ENST00000581819.1 |
RBBP8
|
retinoblastoma binding protein 8 |
| chr11_-_85430163 | 0.05 |
ENST00000529581.1
ENST00000533577.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr10_+_70480963 | 0.05 |
ENST00000265872.6
ENST00000535016.1 ENST00000538031.1 ENST00000543719.1 ENST00000539539.1 ENST00000543225.1 ENST00000536012.1 ENST00000494903.2 |
CCAR1
|
cell division cycle and apoptosis regulator 1 |
| chr1_-_93645818 | 0.05 |
ENST00000370280.1
ENST00000479918.1 |
TMED5
|
transmembrane emp24 protein transport domain containing 5 |
| chr10_+_115511434 | 0.05 |
ENST00000369312.4
|
PLEKHS1
|
pleckstrin homology domain containing, family S member 1 |
| chr4_+_54243862 | 0.05 |
ENST00000306932.6
|
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr17_-_37764128 | 0.05 |
ENST00000302584.4
|
NEUROD2
|
neuronal differentiation 2 |
| chr11_-_76155618 | 0.05 |
ENST00000530759.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr12_-_89919220 | 0.05 |
ENST00000549035.1
ENST00000393179.4 |
POC1B
|
POC1 centriolar protein B |
| chr1_+_172502336 | 0.05 |
ENST00000263688.3
|
SUCO
|
SUN domain containing ossification factor |
| chr13_-_41593425 | 0.05 |
ENST00000239882.3
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chr10_-_33247124 | 0.05 |
ENST00000414670.1
ENST00000302278.3 ENST00000374956.4 ENST00000488494.1 ENST00000417122.2 ENST00000474568.1 |
ITGB1
|
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) |
| chr10_+_115511213 | 0.05 |
ENST00000361048.1
|
PLEKHS1
|
pleckstrin homology domain containing, family S member 1 |
| chr3_-_167813672 | 0.05 |
ENST00000470487.1
|
GOLIM4
|
golgi integral membrane protein 4 |
| chr11_+_125365110 | 0.05 |
ENST00000527818.1
|
AP000708.1
|
AP000708.1 |
| chr7_+_87563557 | 0.05 |
ENST00000439864.1
ENST00000412441.1 ENST00000398201.4 ENST00000265727.7 ENST00000315984.7 ENST00000398209.3 |
ADAM22
|
ADAM metallopeptidase domain 22 |
| chr2_-_39348137 | 0.05 |
ENST00000426016.1
|
SOS1
|
son of sevenless homolog 1 (Drosophila) |
| chr4_+_113152881 | 0.05 |
ENST00000274000.5
|
AP1AR
|
adaptor-related protein complex 1 associated regulatory protein |
| chrX_+_135579238 | 0.05 |
ENST00000535601.1
ENST00000448450.1 ENST00000425695.1 |
HTATSF1
|
HIV-1 Tat specific factor 1 |
| chr3_-_57583052 | 0.05 |
ENST00000496292.1
ENST00000489843.1 |
ARF4
|
ADP-ribosylation factor 4 |
| chr12_-_45270151 | 0.05 |
ENST00000429094.2
|
NELL2
|
NEL-like 2 (chicken) |
| chr15_+_77224045 | 0.05 |
ENST00000320963.5
ENST00000394883.3 |
RCN2
|
reticulocalbin 2, EF-hand calcium binding domain |
| chr2_-_86564740 | 0.04 |
ENST00000540790.1
ENST00000428491.1 |
REEP1
|
receptor accessory protein 1 |
| chr4_+_146560245 | 0.04 |
ENST00000541599.1
|
MMAA
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr4_+_113152978 | 0.04 |
ENST00000309703.6
|
AP1AR
|
adaptor-related protein complex 1 associated regulatory protein |
| chr1_-_198906528 | 0.04 |
ENST00000432296.1
|
MIR181A1HG
|
MIR181A1 host gene (non-protein coding) |
| chr2_+_149402989 | 0.04 |
ENST00000397424.2
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr7_+_134464414 | 0.04 |
ENST00000361901.2
|
CALD1
|
caldesmon 1 |
| chr17_-_56065540 | 0.04 |
ENST00000583932.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr8_-_42358742 | 0.04 |
ENST00000517366.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr4_+_70916119 | 0.04 |
ENST00000246896.3
ENST00000511674.1 |
HTN1
|
histatin 1 |
| chr1_+_172502244 | 0.04 |
ENST00000610051.1
|
SUCO
|
SUN domain containing ossification factor |
| chr17_-_46716647 | 0.04 |
ENST00000608940.1
|
RP11-357H14.17
|
RP11-357H14.17 |
| chr5_-_39425290 | 0.04 |
ENST00000545653.1
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr5_-_39425068 | 0.04 |
ENST00000515700.1
ENST00000339788.6 |
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr7_+_69064566 | 0.04 |
ENST00000403018.2
|
AUTS2
|
autism susceptibility candidate 2 |
| chr10_-_128359074 | 0.04 |
ENST00000544758.1
|
C10orf90
|
chromosome 10 open reading frame 90 |
| chr11_-_85430356 | 0.04 |
ENST00000526999.1
|
SYTL2
|
synaptotagmin-like 2 |
| chrX_+_107068959 | 0.04 |
ENST00000451923.1
|
MID2
|
midline 2 |
| chr7_+_77167343 | 0.04 |
ENST00000433369.2
ENST00000415482.2 |
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr17_+_75447326 | 0.04 |
ENST00000591088.1
|
SEPT9
|
septin 9 |
| chr11_-_82708519 | 0.04 |
ENST00000534301.1
|
RAB30
|
RAB30, member RAS oncogene family |
| chr12_+_69633407 | 0.04 |
ENST00000551516.1
|
CPSF6
|
cleavage and polyadenylation specific factor 6, 68kDa |
| chr5_+_49962772 | 0.04 |
ENST00000281631.5
ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr1_+_200993071 | 0.04 |
ENST00000446333.1
ENST00000458003.1 |
RP11-168O16.1
|
RP11-168O16.1 |
| chr1_+_25071848 | 0.04 |
ENST00000374379.4
|
CLIC4
|
chloride intracellular channel 4 |
| chr14_-_91884150 | 0.04 |
ENST00000553403.1
|
CCDC88C
|
coiled-coil domain containing 88C |
| chr11_-_69519410 | 0.04 |
ENST00000294312.3
|
FGF19
|
fibroblast growth factor 19 |
| chr14_+_103851712 | 0.04 |
ENST00000440884.3
ENST00000416682.2 ENST00000429436.2 ENST00000303622.9 |
MARK3
|
MAP/microtubule affinity-regulating kinase 3 |
| chr2_+_27805880 | 0.04 |
ENST00000379717.1
ENST00000355467.4 ENST00000556601.1 ENST00000416005.2 |
ZNF512
|
zinc finger protein 512 |
| chr3_-_195270162 | 0.04 |
ENST00000438848.1
ENST00000328432.3 |
PPP1R2
|
protein phosphatase 1, regulatory (inhibitor) subunit 2 |
| chr21_+_30671690 | 0.04 |
ENST00000399921.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr1_+_1266654 | 0.04 |
ENST00000339381.5
|
TAS1R3
|
taste receptor, type 1, member 3 |
| chr6_+_13925098 | 0.04 |
ENST00000488300.1
ENST00000544682.1 ENST00000420478.2 |
RNF182
|
ring finger protein 182 |
| chr2_+_27805971 | 0.04 |
ENST00000413371.2
|
ZNF512
|
zinc finger protein 512 |
| chr12_-_45270077 | 0.04 |
ENST00000551601.1
ENST00000549027.1 ENST00000452445.2 |
NELL2
|
NEL-like 2 (chicken) |
| chr4_+_95128996 | 0.04 |
ENST00000457823.2
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr9_-_74979420 | 0.04 |
ENST00000343431.2
ENST00000376956.3 |
ZFAND5
|
zinc finger, AN1-type domain 5 |
| chr4_+_54243798 | 0.04 |
ENST00000337488.6
ENST00000358575.5 ENST00000507922.1 |
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chrX_+_135579670 | 0.04 |
ENST00000218364.4
|
HTATSF1
|
HIV-1 Tat specific factor 1 |
| chr15_+_52043813 | 0.04 |
ENST00000435126.2
|
TMOD2
|
tropomodulin 2 (neuronal) |
| chr20_-_52645231 | 0.03 |
ENST00000448484.1
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr19_-_17571722 | 0.03 |
ENST00000301944.2
|
NXNL1
|
nucleoredoxin-like 1 |
| chr4_+_95129061 | 0.03 |
ENST00000354268.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr2_+_54683419 | 0.03 |
ENST00000356805.4
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1 |
| chr21_-_40720974 | 0.03 |
ENST00000380748.1
|
HMGN1
|
high mobility group nucleosome binding domain 1 |
| chr15_+_93426514 | 0.03 |
ENST00000556722.1
|
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr11_+_34654011 | 0.03 |
ENST00000531794.1
|
EHF
|
ets homologous factor |
| chr1_+_93645314 | 0.03 |
ENST00000343253.7
|
CCDC18
|
coiled-coil domain containing 18 |
| chr5_-_39424961 | 0.03 |
ENST00000503513.1
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr15_+_57211318 | 0.03 |
ENST00000557947.1
|
TCF12
|
transcription factor 12 |
| chr10_-_25241499 | 0.03 |
ENST00000376378.1
ENST00000376376.3 ENST00000320152.6 |
PRTFDC1
|
phosphoribosyl transferase domain containing 1 |
| chr20_-_17511962 | 0.03 |
ENST00000377873.3
|
BFSP1
|
beaded filament structural protein 1, filensin |
| chrM_+_8527 | 0.03 |
ENST00000361899.2
|
MT-ATP6
|
mitochondrially encoded ATP synthase 6 |
| chr19_+_47105309 | 0.03 |
ENST00000599839.1
ENST00000596362.1 |
CALM3
|
calmodulin 3 (phosphorylase kinase, delta) |
| chr7_+_134464376 | 0.03 |
ENST00000454108.1
ENST00000361675.2 |
CALD1
|
caldesmon 1 |
| chr11_-_6677018 | 0.03 |
ENST00000299441.3
|
DCHS1
|
dachsous cadherin-related 1 |
| chr7_+_116312411 | 0.03 |
ENST00000456159.1
ENST00000397752.3 ENST00000318493.6 |
MET
|
met proto-oncogene |
| chr6_-_24877490 | 0.03 |
ENST00000540914.1
ENST00000378023.4 |
FAM65B
|
family with sequence similarity 65, member B |
| chr16_-_31085033 | 0.03 |
ENST00000414399.1
|
ZNF668
|
zinc finger protein 668 |
| chr4_+_71570430 | 0.03 |
ENST00000417478.2
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chrX_+_80457442 | 0.03 |
ENST00000373212.5
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like |
| chr22_+_32439019 | 0.03 |
ENST00000266088.4
|
SLC5A1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr2_-_157198860 | 0.03 |
ENST00000409572.1
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr3_+_52719936 | 0.03 |
ENST00000418458.1
ENST00000394799.2 |
GNL3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr4_+_41614909 | 0.03 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr4_+_55524085 | 0.03 |
ENST00000412167.2
ENST00000288135.5 |
KIT
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog |
| chr8_-_66753682 | 0.03 |
ENST00000396642.3
|
PDE7A
|
phosphodiesterase 7A |
| chr9_-_98269481 | 0.03 |
ENST00000418258.1
ENST00000553011.1 ENST00000551845.1 |
PTCH1
|
patched 1 |
| chr18_+_55712915 | 0.03 |
ENST00000592846.1
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr15_+_74610894 | 0.03 |
ENST00000558821.1
ENST00000268082.4 |
CCDC33
|
coiled-coil domain containing 33 |
| chr19_+_35783047 | 0.03 |
ENST00000595791.1
ENST00000597035.1 ENST00000537831.2 |
MAG
|
myelin associated glycoprotein |
| chr9_-_98268883 | 0.03 |
ENST00000551630.1
ENST00000548420.1 |
PTCH1
|
patched 1 |
| chr4_+_41614720 | 0.03 |
ENST00000509277.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr18_+_7946941 | 0.03 |
ENST00000444013.1
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M |
| chr1_+_212797789 | 0.03 |
ENST00000294829.3
|
FAM71A
|
family with sequence similarity 71, member A |
| chr17_+_67498538 | 0.03 |
ENST00000589647.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr14_-_38036271 | 0.03 |
ENST00000556024.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr6_-_3227877 | 0.03 |
ENST00000259818.7
|
TUBB2B
|
tubulin, beta 2B class IIb |
| chr8_-_30515693 | 0.03 |
ENST00000355904.4
|
GTF2E2
|
general transcription factor IIE, polypeptide 2, beta 34kDa |
| chr13_-_24007815 | 0.03 |
ENST00000382298.3
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr9_+_125795788 | 0.03 |
ENST00000373643.5
|
RABGAP1
|
RAB GTPase activating protein 1 |
| chr5_+_139175380 | 0.03 |
ENST00000274710.3
|
PSD2
|
pleckstrin and Sec7 domain containing 2 |
| chr16_-_22385901 | 0.03 |
ENST00000268383.2
|
CDR2
|
cerebellar degeneration-related protein 2, 62kDa |
| chr21_-_16437126 | 0.03 |
ENST00000318948.4
|
NRIP1
|
nuclear receptor interacting protein 1 |
| chr4_-_52883786 | 0.03 |
ENST00000343457.3
|
LRRC66
|
leucine rich repeat containing 66 |
| chr19_+_35783037 | 0.03 |
ENST00000361922.4
|
MAG
|
myelin associated glycoprotein |
| chr11_-_130184470 | 0.03 |
ENST00000357899.4
ENST00000397753.1 |
ZBTB44
|
zinc finger and BTB domain containing 44 |
| chr6_+_64282447 | 0.03 |
ENST00000370650.2
ENST00000578299.1 |
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr2_-_70475701 | 0.03 |
ENST00000282574.4
|
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr4_-_71705027 | 0.03 |
ENST00000545193.1
|
GRSF1
|
G-rich RNA sequence binding factor 1 |
| chr19_+_45204630 | 0.03 |
ENST00000405314.2
|
CEACAM16
|
carcinoembryonic antigen-related cell adhesion molecule 16 |
| chr17_-_76719638 | 0.03 |
ENST00000587308.1
|
CYTH1
|
cytohesin 1 |
| chr4_-_71705060 | 0.03 |
ENST00000514161.1
|
GRSF1
|
G-rich RNA sequence binding factor 1 |
| chr2_+_149402553 | 0.03 |
ENST00000258484.6
ENST00000409654.1 |
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr9_+_108463234 | 0.03 |
ENST00000374688.1
|
TMEM38B
|
transmembrane protein 38B |
| chr12_-_105478339 | 0.03 |
ENST00000424857.2
ENST00000258494.9 |
ALDH1L2
|
aldehyde dehydrogenase 1 family, member L2 |
| chr19_+_35783028 | 0.03 |
ENST00000600291.1
ENST00000392213.3 |
MAG
|
myelin associated glycoprotein |
| chr6_+_4773205 | 0.02 |
ENST00000440139.1
|
CDYL
|
chromodomain protein, Y-like |
| chr1_-_182360498 | 0.02 |
ENST00000417584.2
|
GLUL
|
glutamate-ammonia ligase |
| chr1_-_89357179 | 0.02 |
ENST00000448623.1
ENST00000418217.1 ENST00000370500.5 |
GTF2B
|
general transcription factor IIB |
| chr14_-_55878538 | 0.02 |
ENST00000247178.5
|
ATG14
|
autophagy related 14 |
| chr17_+_30813576 | 0.02 |
ENST00000313401.3
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
| chr11_-_64013663 | 0.02 |
ENST00000392210.2
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr21_+_35552978 | 0.02 |
ENST00000428914.2
ENST00000609062.1 ENST00000609947.1 |
LINC00310
|
long intergenic non-protein coding RNA 310 |
| chr15_-_56035177 | 0.02 |
ENST00000389286.4
ENST00000561292.1 |
PRTG
|
protogenin |
| chr6_-_31782813 | 0.02 |
ENST00000375654.4
|
HSPA1L
|
heat shock 70kDa protein 1-like |
| chr7_+_73868439 | 0.02 |
ENST00000424337.2
|
GTF2IRD1
|
GTF2I repeat domain containing 1 |
| chr1_+_182808474 | 0.02 |
ENST00000367549.3
|
DHX9
|
DEAH (Asp-Glu-Ala-His) box helicase 9 |
| chr14_+_23305760 | 0.02 |
ENST00000311852.6
|
MMP14
|
matrix metallopeptidase 14 (membrane-inserted) |
| chr16_-_15736881 | 0.02 |
ENST00000540441.2
|
KIAA0430
|
KIAA0430 |
| chr4_+_88896819 | 0.02 |
ENST00000237623.7
ENST00000395080.3 ENST00000508233.1 ENST00000360804.4 |
SPP1
|
secreted phosphoprotein 1 |
| chr7_-_50628745 | 0.02 |
ENST00000380984.4
ENST00000357936.5 ENST00000426377.1 |
DDC
|
dopa decarboxylase (aromatic L-amino acid decarboxylase) |
| chr4_-_68620053 | 0.02 |
ENST00000420975.2
ENST00000226413.4 |
GNRHR
|
gonadotropin-releasing hormone receptor |
| chr4_+_71588372 | 0.02 |
ENST00000536664.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr5_-_176900610 | 0.02 |
ENST00000477391.2
ENST00000393565.1 ENST00000309007.5 |
DBN1
|
drebrin 1 |
| chr13_-_108518986 | 0.02 |
ENST00000375915.2
|
FAM155A
|
family with sequence similarity 155, member A |
| chr1_+_24286287 | 0.02 |
ENST00000334351.7
ENST00000374468.1 |
PNRC2
|
proline-rich nuclear receptor coactivator 2 |
| chr22_+_29168652 | 0.02 |
ENST00000249064.4
ENST00000444523.1 ENST00000448492.2 ENST00000421503.2 |
CCDC117
|
coiled-coil domain containing 117 |
| chr2_-_204399976 | 0.02 |
ENST00000457812.1
|
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr14_-_35344093 | 0.02 |
ENST00000382422.2
|
BAZ1A
|
bromodomain adjacent to zinc finger domain, 1A |
| chr1_+_150039369 | 0.02 |
ENST00000369130.3
ENST00000369128.5 |
VPS45
|
vacuolar protein sorting 45 homolog (S. cerevisiae) |
| chr10_-_32667660 | 0.02 |
ENST00000375110.2
|
EPC1
|
enhancer of polycomb homolog 1 (Drosophila) |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.1 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.0 | 0.2 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.1 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 | 0.1 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.1 | GO:0051086 | chaperone mediated protein folding independent of cofactor(GO:0051086) |
| 0.0 | 0.2 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.0 | 0.0 | GO:0001835 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.0 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.0 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.0 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 | 0.0 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.0 | GO:0051866 | general adaptation syndrome(GO:0051866) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097453 | mesaxon(GO:0097453) ensheathing process(GO:1990015) |
| 0.0 | 0.0 | GO:0034677 | integrin alpha7-beta1 complex(GO:0034677) |
| 0.0 | 0.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 0.0 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |