Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
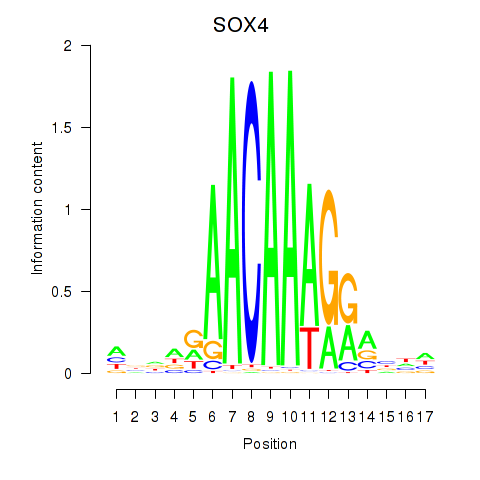
Results for SOX4
Z-value: 0.83
Transcription factors associated with SOX4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX4
|
ENSG00000124766.4 | SRY-box transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX4 | hg19_v2_chr6_+_21593972_21594071 | 1.00 | 4.3e-04 | Click! |
Activity profile of SOX4 motif
Sorted Z-values of SOX4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_23791484 | 1.12 |
ENST00000594872.1
|
AL049829.1
|
Uncharacterized protein |
| chr5_+_102200948 | 0.63 |
ENST00000511477.1
ENST00000506006.1 ENST00000509832.1 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr13_+_110958124 | 0.48 |
ENST00000400163.2
|
COL4A2
|
collagen, type IV, alpha 2 |
| chr22_-_42336209 | 0.48 |
ENST00000472374.2
|
CENPM
|
centromere protein M |
| chr15_+_91416092 | 0.47 |
ENST00000559353.1
|
FURIN
|
furin (paired basic amino acid cleaving enzyme) |
| chr2_-_118943930 | 0.45 |
ENST00000449075.1
ENST00000414886.1 ENST00000449819.1 |
AC093901.1
|
AC093901.1 |
| chr22_-_21984282 | 0.42 |
ENST00000398873.3
ENST00000292778.6 |
YDJC
|
YdjC homolog (bacterial) |
| chr4_-_75024085 | 0.42 |
ENST00000600169.1
|
AC093677.1
|
Uncharacterized protein |
| chr19_-_59030921 | 0.42 |
ENST00000354590.3
ENST00000596739.1 |
ZBTB45
|
zinc finger and BTB domain containing 45 |
| chr19_+_56111680 | 0.38 |
ENST00000301073.3
|
ZNF524
|
zinc finger protein 524 |
| chr7_-_105926058 | 0.36 |
ENST00000417537.1
|
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chr11_-_10830463 | 0.36 |
ENST00000527419.1
ENST00000530211.1 ENST00000530702.1 ENST00000524932.1 ENST00000532570.1 |
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr19_-_59031118 | 0.36 |
ENST00000600990.1
|
ZBTB45
|
zinc finger and BTB domain containing 45 |
| chr2_+_201981663 | 0.33 |
ENST00000433445.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr17_+_67498396 | 0.33 |
ENST00000588110.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr12_-_66317967 | 0.33 |
ENST00000601398.1
|
AC090673.2
|
Uncharacterized protein |
| chr14_-_20922960 | 0.32 |
ENST00000553640.1
ENST00000488532.2 |
OSGEP
|
O-sialoglycoprotein endopeptidase |
| chr3_+_181429704 | 0.31 |
ENST00000431565.2
ENST00000325404.1 |
SOX2
|
SRY (sex determining region Y)-box 2 |
| chr2_-_175202151 | 0.30 |
ENST00000595354.1
|
AC018470.1
|
Uncharacterized protein FLJ46347 |
| chr15_-_74494779 | 0.30 |
ENST00000571341.1
|
STRA6
|
stimulated by retinoic acid 6 |
| chr7_+_150076406 | 0.30 |
ENST00000329630.5
|
ZNF775
|
zinc finger protein 775 |
| chr15_-_41166414 | 0.28 |
ENST00000220507.4
|
RHOV
|
ras homolog family member V |
| chr18_+_3450161 | 0.27 |
ENST00000551402.1
ENST00000577543.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr13_+_111855399 | 0.27 |
ENST00000426768.2
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr9_-_131940526 | 0.26 |
ENST00000372491.2
|
IER5L
|
immediate early response 5-like |
| chr7_-_84121858 | 0.26 |
ENST00000448879.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr3_-_134092561 | 0.25 |
ENST00000510560.1
ENST00000504234.1 ENST00000515172.1 |
AMOTL2
|
angiomotin like 2 |
| chr10_-_18948156 | 0.25 |
ENST00000414939.1
ENST00000449529.1 ENST00000456217.1 ENST00000444660.1 |
ARL5B-AS1
|
ARL5B antisense RNA 1 |
| chr1_-_243418650 | 0.25 |
ENST00000522995.1
|
CEP170
|
centrosomal protein 170kDa |
| chr7_-_111032971 | 0.24 |
ENST00000450877.1
|
IMMP2L
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr22_+_37959647 | 0.24 |
ENST00000415670.1
|
CDC42EP1
|
CDC42 effector protein (Rho GTPase binding) 1 |
| chr7_+_99699179 | 0.24 |
ENST00000438383.1
ENST00000429084.1 ENST00000359593.4 ENST00000439416.1 |
AP4M1
|
adaptor-related protein complex 4, mu 1 subunit |
| chr1_-_1243392 | 0.23 |
ENST00000354700.5
|
ACAP3
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3 |
| chr6_-_127840048 | 0.23 |
ENST00000467753.1
|
SOGA3
|
SOGA family member 3 |
| chr12_-_76879852 | 0.22 |
ENST00000548341.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chrX_-_2847366 | 0.22 |
ENST00000381154.1
|
ARSD
|
arylsulfatase D |
| chr7_-_27219849 | 0.22 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr1_-_153940097 | 0.21 |
ENST00000413622.1
ENST00000310483.6 |
SLC39A1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr14_+_69658480 | 0.20 |
ENST00000409949.1
ENST00000409242.1 ENST00000312994.5 ENST00000413191.1 |
EXD2
|
exonuclease 3'-5' domain containing 2 |
| chr16_+_30406423 | 0.20 |
ENST00000524644.1
|
ZNF48
|
zinc finger protein 48 |
| chr7_+_99699280 | 0.20 |
ENST00000421755.1
|
AP4M1
|
adaptor-related protein complex 4, mu 1 subunit |
| chr16_-_73082274 | 0.19 |
ENST00000268489.5
|
ZFHX3
|
zinc finger homeobox 3 |
| chr11_-_62414070 | 0.19 |
ENST00000540933.1
ENST00000346178.4 ENST00000356638.3 ENST00000534779.1 ENST00000525994.1 |
GANAB
|
glucosidase, alpha; neutral AB |
| chr19_+_49128209 | 0.19 |
ENST00000599748.1
ENST00000443164.1 ENST00000599029.1 |
SPHK2
|
sphingosine kinase 2 |
| chr2_-_20425158 | 0.18 |
ENST00000381150.1
|
SDC1
|
syndecan 1 |
| chr4_-_170192000 | 0.18 |
ENST00000502315.1
|
SH3RF1
|
SH3 domain containing ring finger 1 |
| chr15_+_81293254 | 0.18 |
ENST00000267984.2
|
MESDC1
|
mesoderm development candidate 1 |
| chr5_-_175964366 | 0.18 |
ENST00000274811.4
|
RNF44
|
ring finger protein 44 |
| chr5_-_146833803 | 0.18 |
ENST00000512722.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr19_+_1249869 | 0.18 |
ENST00000591446.2
|
MIDN
|
midnolin |
| chr1_-_1243252 | 0.17 |
ENST00000353662.3
|
ACAP3
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3 |
| chr20_-_52612705 | 0.17 |
ENST00000434986.2
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr1_-_152297679 | 0.16 |
ENST00000368799.1
|
FLG
|
filaggrin |
| chr14_-_61124977 | 0.16 |
ENST00000554986.1
|
SIX1
|
SIX homeobox 1 |
| chr7_+_134551583 | 0.16 |
ENST00000435928.1
|
CALD1
|
caldesmon 1 |
| chr1_+_153940713 | 0.16 |
ENST00000368601.1
ENST00000368603.1 ENST00000368600.3 |
CREB3L4
|
cAMP responsive element binding protein 3-like 4 |
| chr7_-_35293740 | 0.16 |
ENST00000408931.3
|
TBX20
|
T-box 20 |
| chr17_+_67498295 | 0.16 |
ENST00000589295.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr19_+_1248547 | 0.16 |
ENST00000586757.1
ENST00000300952.2 |
MIDN
|
midnolin |
| chr2_+_105050794 | 0.16 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr15_+_75940218 | 0.16 |
ENST00000308527.5
|
SNX33
|
sorting nexin 33 |
| chr11_+_5711010 | 0.15 |
ENST00000454828.1
|
TRIM22
|
tripartite motif containing 22 |
| chr5_+_140739537 | 0.15 |
ENST00000522605.1
|
PCDHGB2
|
protocadherin gamma subfamily B, 2 |
| chr5_+_140579162 | 0.15 |
ENST00000536699.1
ENST00000354757.3 |
PCDHB11
|
protocadherin beta 11 |
| chr15_-_70390213 | 0.15 |
ENST00000557997.1
ENST00000317509.8 ENST00000442299.2 |
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chr11_-_71810258 | 0.15 |
ENST00000544594.1
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 |
| chr12_+_95611536 | 0.15 |
ENST00000549002.1
|
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr9_+_139221880 | 0.15 |
ENST00000392945.3
ENST00000440944.1 |
GPSM1
|
G-protein signaling modulator 1 |
| chr15_-_74495188 | 0.15 |
ENST00000563965.1
ENST00000395105.4 |
STRA6
|
stimulated by retinoic acid 6 |
| chr1_-_85930246 | 0.14 |
ENST00000426972.3
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr8_+_61822605 | 0.14 |
ENST00000526936.1
|
AC022182.1
|
AC022182.1 |
| chr3_-_171489085 | 0.14 |
ENST00000418087.1
|
PLD1
|
phospholipase D1, phosphatidylcholine-specific |
| chr7_-_150946015 | 0.14 |
ENST00000262188.8
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr12_+_132379160 | 0.13 |
ENST00000321867.4
|
ULK1
|
unc-51 like autophagy activating kinase 1 |
| chr1_+_44870866 | 0.13 |
ENST00000355387.2
ENST00000361799.2 |
RNF220
|
ring finger protein 220 |
| chr8_-_6420759 | 0.13 |
ENST00000523120.1
|
ANGPT2
|
angiopoietin 2 |
| chr14_+_24439148 | 0.13 |
ENST00000543805.1
ENST00000534993.1 |
DHRS4L2
|
dehydrogenase/reductase (SDR family) member 4 like 2 |
| chr2_-_85641162 | 0.13 |
ENST00000447219.2
ENST00000409670.1 ENST00000409724.1 |
CAPG
|
capping protein (actin filament), gelsolin-like |
| chrX_-_24690771 | 0.12 |
ENST00000379145.1
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr15_+_25200108 | 0.12 |
ENST00000577949.1
ENST00000338094.6 ENST00000338327.4 ENST00000579070.1 ENST00000577565.1 |
SNURF
SNRPN
|
SNRPN upstream reading frame protein small nuclear ribonucleoprotein polypeptide N |
| chr14_+_50234827 | 0.12 |
ENST00000554589.1
ENST00000557247.1 |
KLHDC2
|
kelch domain containing 2 |
| chr1_-_161015663 | 0.12 |
ENST00000534633.1
|
USF1
|
upstream transcription factor 1 |
| chr14_+_101299520 | 0.12 |
ENST00000455531.1
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chrX_-_47930980 | 0.12 |
ENST00000442455.3
ENST00000428686.1 ENST00000276054.4 |
ZNF630
|
zinc finger protein 630 |
| chr4_-_40516560 | 0.11 |
ENST00000513473.1
|
RBM47
|
RNA binding motif protein 47 |
| chr22_-_31688431 | 0.11 |
ENST00000402249.3
ENST00000443175.1 ENST00000215912.5 ENST00000441972.1 |
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chrX_+_9433289 | 0.11 |
ENST00000422314.1
|
TBL1X
|
transducin (beta)-like 1X-linked |
| chr7_+_23146271 | 0.11 |
ENST00000545771.1
|
KLHL7
|
kelch-like family member 7 |
| chr14_+_91580357 | 0.11 |
ENST00000298858.4
ENST00000521081.1 ENST00000520328.1 ENST00000256324.10 ENST00000524232.1 ENST00000522170.1 ENST00000519950.1 ENST00000523879.1 ENST00000521077.2 ENST00000518665.2 |
C14orf159
|
chromosome 14 open reading frame 159 |
| chr12_-_62997214 | 0.11 |
ENST00000408887.2
|
C12orf61
|
chromosome 12 open reading frame 61 |
| chrX_-_73512411 | 0.11 |
ENST00000602576.1
ENST00000429124.1 |
FTX
|
FTX transcript, XIST regulator (non-protein coding) |
| chr12_+_28605426 | 0.11 |
ENST00000542801.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr1_+_153940741 | 0.10 |
ENST00000431292.1
|
CREB3L4
|
cAMP responsive element binding protein 3-like 4 |
| chr20_-_52612468 | 0.10 |
ENST00000422805.1
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr1_+_153940346 | 0.10 |
ENST00000405694.3
ENST00000449724.1 ENST00000368607.3 ENST00000271889.4 |
CREB3L4
|
cAMP responsive element binding protein 3-like 4 |
| chr17_+_26698677 | 0.09 |
ENST00000457710.3
|
SARM1
|
sterile alpha and TIR motif containing 1 |
| chr6_-_127840021 | 0.09 |
ENST00000465909.2
|
SOGA3
|
SOGA family member 3 |
| chr5_+_86563636 | 0.09 |
ENST00000274376.6
|
RASA1
|
RAS p21 protein activator (GTPase activating protein) 1 |
| chr9_-_117150303 | 0.09 |
ENST00000312033.3
|
AKNA
|
AT-hook transcription factor |
| chr2_-_114647327 | 0.09 |
ENST00000602760.1
|
RP11-141B14.1
|
RP11-141B14.1 |
| chr20_-_35492048 | 0.09 |
ENST00000237536.4
|
SOGA1
|
suppressor of glucose, autophagy associated 1 |
| chr7_+_141408153 | 0.09 |
ENST00000397541.2
|
WEE2
|
WEE1 homolog 2 (S. pombe) |
| chr15_+_25200074 | 0.09 |
ENST00000390687.4
ENST00000584968.1 ENST00000346403.6 ENST00000554227.2 |
SNRPN
|
small nuclear ribonucleoprotein polypeptide N |
| chr3_-_38992052 | 0.09 |
ENST00000302328.3
ENST00000450244.1 ENST00000444237.2 |
SCN11A
|
sodium channel, voltage-gated, type XI, alpha subunit |
| chr7_-_148725544 | 0.09 |
ENST00000413966.1
|
PDIA4
|
protein disulfide isomerase family A, member 4 |
| chr17_-_14683517 | 0.08 |
ENST00000379640.1
|
AC005863.1
|
AC005863.1 |
| chr11_+_100784231 | 0.08 |
ENST00000531183.1
|
ARHGAP42
|
Rho GTPase activating protein 42 |
| chrX_+_12993202 | 0.08 |
ENST00000451311.2
ENST00000380636.1 |
TMSB4X
|
thymosin beta 4, X-linked |
| chrX_-_73512358 | 0.08 |
ENST00000602776.1
|
FTX
|
FTX transcript, XIST regulator (non-protein coding) |
| chr15_-_70390191 | 0.08 |
ENST00000559191.1
|
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chr17_+_80416050 | 0.08 |
ENST00000579198.1
ENST00000390006.4 ENST00000580296.1 |
NARF
|
nuclear prelamin A recognition factor |
| chr17_+_27920486 | 0.08 |
ENST00000394859.3
|
ANKRD13B
|
ankyrin repeat domain 13B |
| chrX_-_153599578 | 0.08 |
ENST00000360319.4
ENST00000344736.4 |
FLNA
|
filamin A, alpha |
| chr1_+_209602771 | 0.08 |
ENST00000440276.1
|
MIR205HG
|
MIR205 host gene (non-protein coding) |
| chr12_-_122296755 | 0.08 |
ENST00000289004.4
|
HPD
|
4-hydroxyphenylpyruvate dioxygenase |
| chr1_-_108231101 | 0.08 |
ENST00000544443.1
ENST00000415432.2 |
VAV3
|
vav 3 guanine nucleotide exchange factor |
| chr15_-_34502197 | 0.08 |
ENST00000557877.1
|
KATNBL1
|
katanin p80 subunit B-like 1 |
| chr5_+_140749803 | 0.07 |
ENST00000576222.1
|
PCDHGB3
|
protocadherin gamma subfamily B, 3 |
| chr10_+_24755416 | 0.07 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr13_+_39612442 | 0.07 |
ENST00000470258.1
ENST00000379600.3 |
NHLRC3
|
NHL repeat containing 3 |
| chr10_+_22605304 | 0.07 |
ENST00000475460.2
ENST00000602390.1 ENST00000489125.2 ENST00000456711.1 ENST00000444869.1 |
COMMD3-BMI1
COMMD3
|
COMMD3-BMI1 readthrough COMM domain containing 3 |
| chr16_+_524900 | 0.07 |
ENST00000449879.1
|
RAB11FIP3
|
RAB11 family interacting protein 3 (class II) |
| chr16_+_30406721 | 0.07 |
ENST00000320159.2
|
ZNF48
|
zinc finger protein 48 |
| chr17_+_67498538 | 0.07 |
ENST00000589647.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr2_-_70995307 | 0.07 |
ENST00000264436.4
ENST00000355733.3 ENST00000447731.2 ENST00000430656.1 ENST00000413157.2 |
ADD2
|
adducin 2 (beta) |
| chr7_+_20370300 | 0.07 |
ENST00000537992.1
|
ITGB8
|
integrin, beta 8 |
| chr7_-_84122033 | 0.07 |
ENST00000424555.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr5_+_140235469 | 0.06 |
ENST00000506939.2
ENST00000307360.5 |
PCDHA10
|
protocadherin alpha 10 |
| chr1_-_161015752 | 0.06 |
ENST00000435396.1
ENST00000368021.3 |
USF1
|
upstream transcription factor 1 |
| chr7_-_27219632 | 0.06 |
ENST00000470747.4
|
RP1-170O19.20
|
Uncharacterized protein |
| chr10_+_11206925 | 0.06 |
ENST00000354440.2
ENST00000315874.4 ENST00000427450.1 |
CELF2
|
CUGBP, Elav-like family member 2 |
| chr2_-_37501692 | 0.06 |
ENST00000443977.1
|
PRKD3
|
protein kinase D3 |
| chr4_-_22444733 | 0.06 |
ENST00000508133.1
|
GPR125
|
G protein-coupled receptor 125 |
| chr16_-_48281444 | 0.06 |
ENST00000537808.1
ENST00000569991.1 |
ABCC11
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 11 |
| chr8_-_116504448 | 0.06 |
ENST00000518018.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr5_-_76788298 | 0.06 |
ENST00000511036.1
|
WDR41
|
WD repeat domain 41 |
| chr11_-_94965667 | 0.06 |
ENST00000542176.1
ENST00000278499.2 |
SESN3
|
sestrin 3 |
| chr16_-_31085033 | 0.06 |
ENST00000414399.1
|
ZNF668
|
zinc finger protein 668 |
| chr12_+_109915179 | 0.06 |
ENST00000434735.2
|
UBE3B
|
ubiquitin protein ligase E3B |
| chr16_+_524850 | 0.06 |
ENST00000450428.1
ENST00000452814.1 |
RAB11FIP3
|
RAB11 family interacting protein 3 (class II) |
| chr4_-_159080806 | 0.06 |
ENST00000590648.1
|
FAM198B
|
family with sequence similarity 198, member B |
| chr14_-_71276211 | 0.06 |
ENST00000381250.4
ENST00000555993.2 |
MAP3K9
|
mitogen-activated protein kinase kinase kinase 9 |
| chr19_-_17375541 | 0.06 |
ENST00000252597.3
|
USHBP1
|
Usher syndrome 1C binding protein 1 |
| chrX_+_51927919 | 0.06 |
ENST00000416960.1
|
MAGED4
|
melanoma antigen family D, 4 |
| chr12_-_109915098 | 0.06 |
ENST00000542858.1
ENST00000542262.1 ENST00000424763.2 |
KCTD10
|
potassium channel tetramerization domain containing 10 |
| chr11_-_61560053 | 0.06 |
ENST00000537328.1
|
TMEM258
|
transmembrane protein 258 |
| chr8_-_42358742 | 0.06 |
ENST00000517366.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr11_-_85430356 | 0.05 |
ENST00000526999.1
|
SYTL2
|
synaptotagmin-like 2 |
| chr1_-_32801825 | 0.05 |
ENST00000329421.7
|
MARCKSL1
|
MARCKS-like 1 |
| chr5_+_140787600 | 0.05 |
ENST00000520790.1
|
PCDHGB6
|
protocadherin gamma subfamily B, 6 |
| chrX_-_110655391 | 0.05 |
ENST00000356915.2
ENST00000356220.3 |
DCX
|
doublecortin |
| chr10_-_61900762 | 0.05 |
ENST00000355288.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr1_+_40840320 | 0.05 |
ENST00000372708.1
|
SMAP2
|
small ArfGAP2 |
| chr3_+_50649302 | 0.05 |
ENST00000446044.1
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr22_+_31477296 | 0.05 |
ENST00000426927.1
ENST00000440425.1 ENST00000358743.1 ENST00000347557.2 ENST00000333137.7 |
SMTN
|
smoothelin |
| chr3_-_8686479 | 0.05 |
ENST00000544814.1
ENST00000427408.1 |
SSUH2
|
ssu-2 homolog (C. elegans) |
| chr1_+_28844778 | 0.05 |
ENST00000411533.1
|
RCC1
|
regulator of chromosome condensation 1 |
| chr8_-_8751068 | 0.05 |
ENST00000276282.6
|
MFHAS1
|
malignant fibrous histiocytoma amplified sequence 1 |
| chr13_-_110959478 | 0.05 |
ENST00000543140.1
ENST00000375820.4 |
COL4A1
|
collagen, type IV, alpha 1 |
| chr18_+_7946839 | 0.04 |
ENST00000578916.1
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M |
| chr4_+_118955500 | 0.04 |
ENST00000296499.5
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr15_-_72564906 | 0.04 |
ENST00000566844.1
|
PARP6
|
poly (ADP-ribose) polymerase family, member 6 |
| chr14_+_32798462 | 0.04 |
ENST00000280979.4
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr2_-_165697717 | 0.04 |
ENST00000439313.1
|
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr5_+_140767452 | 0.04 |
ENST00000519479.1
|
PCDHGB4
|
protocadherin gamma subfamily B, 4 |
| chr16_-_48281305 | 0.04 |
ENST00000356608.2
|
ABCC11
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 11 |
| chr7_-_92855762 | 0.04 |
ENST00000453812.2
ENST00000394468.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr8_+_24151553 | 0.04 |
ENST00000265769.4
ENST00000540823.1 ENST00000397649.3 |
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr15_+_78730622 | 0.04 |
ENST00000560440.1
|
IREB2
|
iron-responsive element binding protein 2 |
| chr16_+_21608525 | 0.04 |
ENST00000567404.1
|
METTL9
|
methyltransferase like 9 |
| chr16_-_68482440 | 0.04 |
ENST00000219334.5
|
SMPD3
|
sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II) |
| chr5_-_173043591 | 0.04 |
ENST00000285908.5
ENST00000480951.1 ENST00000311086.4 |
BOD1
|
biorientation of chromosomes in cell division 1 |
| chr4_-_139163491 | 0.04 |
ENST00000280612.5
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chr9_-_39239171 | 0.04 |
ENST00000358144.2
|
CNTNAP3
|
contactin associated protein-like 3 |
| chr20_-_56284816 | 0.04 |
ENST00000395819.3
ENST00000341744.3 |
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr7_-_107643567 | 0.04 |
ENST00000393559.1
ENST00000439976.1 ENST00000393560.1 |
LAMB1
|
laminin, beta 1 |
| chr11_+_125496619 | 0.04 |
ENST00000532669.1
ENST00000278916.3 |
CHEK1
|
checkpoint kinase 1 |
| chr15_-_45670924 | 0.04 |
ENST00000396659.3
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr6_-_64029879 | 0.04 |
ENST00000370658.5
ENST00000485906.2 ENST00000370657.4 |
LGSN
|
lengsin, lens protein with glutamine synthetase domain |
| chr1_-_155880672 | 0.04 |
ENST00000609492.1
ENST00000368322.3 |
RIT1
|
Ras-like without CAAX 1 |
| chrX_-_128977364 | 0.04 |
ENST00000371064.3
|
ZDHHC9
|
zinc finger, DHHC-type containing 9 |
| chr6_+_139456226 | 0.04 |
ENST00000367658.2
|
HECA
|
headcase homolog (Drosophila) |
| chr17_-_38928414 | 0.04 |
ENST00000335552.4
|
KRT26
|
keratin 26 |
| chr4_-_69111401 | 0.03 |
ENST00000332644.5
|
TMPRSS11B
|
transmembrane protease, serine 11B |
| chr14_+_69658194 | 0.03 |
ENST00000409018.3
ENST00000409014.1 ENST00000409675.1 |
EXD2
|
exonuclease 3'-5' domain containing 2 |
| chr1_+_97188188 | 0.03 |
ENST00000541987.1
|
PTBP2
|
polypyrimidine tract binding protein 2 |
| chr2_+_11679963 | 0.03 |
ENST00000263834.5
|
GREB1
|
growth regulation by estrogen in breast cancer 1 |
| chr3_+_111260980 | 0.03 |
ENST00000438817.2
|
CD96
|
CD96 molecule |
| chr14_+_91580732 | 0.03 |
ENST00000519019.1
ENST00000523816.1 ENST00000517518.1 |
C14orf159
|
chromosome 14 open reading frame 159 |
| chr10_-_105845536 | 0.03 |
ENST00000393211.3
|
COL17A1
|
collagen, type XVII, alpha 1 |
| chr10_-_62332357 | 0.03 |
ENST00000503366.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr19_+_3880581 | 0.03 |
ENST00000450849.2
ENST00000301260.6 ENST00000398448.3 |
ATCAY
|
ataxia, cerebellar, Cayman type |
| chr7_-_92146729 | 0.03 |
ENST00000541751.1
|
PEX1
|
peroxisomal biogenesis factor 1 |
| chr12_+_8666126 | 0.03 |
ENST00000299665.2
|
CLEC4D
|
C-type lectin domain family 4, member D |
| chr14_-_73493825 | 0.03 |
ENST00000318876.5
ENST00000556143.1 |
ZFYVE1
|
zinc finger, FYVE domain containing 1 |
| chr5_+_140480083 | 0.03 |
ENST00000231130.2
|
PCDHB3
|
protocadherin beta 3 |
| chr21_+_30672433 | 0.03 |
ENST00000451655.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr9_+_112852477 | 0.03 |
ENST00000480388.1
|
AKAP2
|
A kinase (PRKA) anchor protein 2 |
| chr6_-_139613269 | 0.03 |
ENST00000358430.3
|
TXLNB
|
taxilin beta |
| chr15_-_76304731 | 0.02 |
ENST00000394907.3
|
NRG4
|
neuregulin 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0090472 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.1 | 0.6 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.1 | 0.6 | GO:0018032 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 0.1 | 0.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.3 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.1 | 0.4 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.1 | 0.3 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.2 | GO:0048627 | myoblast development(GO:0048627) |
| 0.1 | 0.3 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 0.2 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.1 | 0.2 | GO:0060620 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.0 | 0.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.2 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 | 0.2 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.0 | 0.1 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.0 | 0.2 | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 | 0.1 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.0 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.0 | 0.3 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.0 | 0.3 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.1 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.0 | 0.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.5 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.2 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.4 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.0 | GO:0030323 | respiratory tube development(GO:0030323) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.1 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.0 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.0 | 0.4 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.1 | GO:0072658 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.1 | GO:0075044 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.0 | 0.2 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.1 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.5 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 0.5 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 0.2 | GO:0036457 | keratohyalin granule(GO:0036457) |
| 0.0 | 0.4 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.2 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.3 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.1 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.3 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 0.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.1 | 0.6 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.1 | 0.3 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.1 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.0 | 0.5 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.2 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.2 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.2 | GO:0047820 | D-glutamate cyclase activity(GO:0047820) |
| 0.0 | 0.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.7 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.0 | GO:0015067 | amidinotransferase activity(GO:0015067) glycine amidinotransferase activity(GO:0015068) |
| 0.0 | 0.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.0 | GO:0043849 | Ras palmitoyltransferase activity(GO:0043849) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.3 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.4 | GO:0051183 | vitamin transporter activity(GO:0051183) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.6 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.5 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |