Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
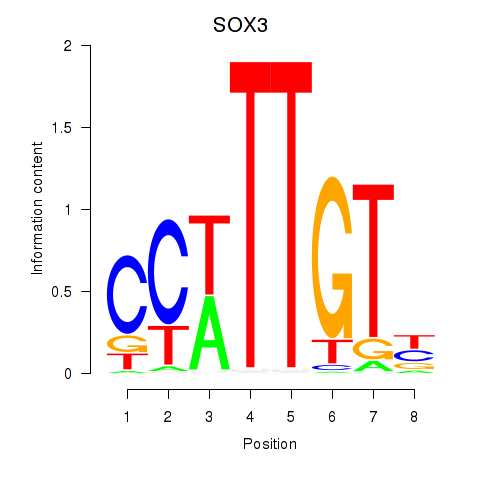
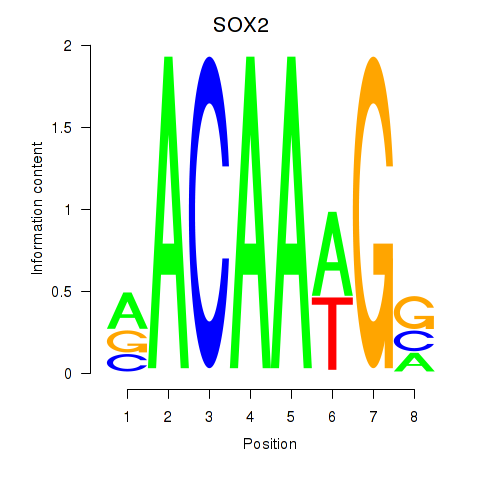
Results for SOX3_SOX2
Z-value: 0.69
Transcription factors associated with SOX3_SOX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX3
|
ENSG00000134595.6 | SRY-box transcription factor 3 |
|
SOX2
|
ENSG00000181449.2 | SRY-box transcription factor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX2 | hg19_v2_chr3_+_181429704_181429722 | -0.46 | 5.4e-01 | Click! |
Activity profile of SOX3_SOX2 motif
Sorted Z-values of SOX3_SOX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_26322987 | 0.33 |
ENST00000505958.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr13_+_111365602 | 0.32 |
ENST00000333219.7
|
ING1
|
inhibitor of growth family, member 1 |
| chr15_+_101402041 | 0.30 |
ENST00000558475.1
ENST00000558641.1 ENST00000559673.1 |
RP11-66B24.1
|
RP11-66B24.1 |
| chr5_-_65018834 | 0.30 |
ENST00000506816.1
|
SGTB
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr10_-_18948208 | 0.29 |
ENST00000607346.1
|
ARL5B-AS1
|
ARL5B antisense RNA 1 |
| chr14_-_51027838 | 0.29 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr12_+_95611536 | 0.28 |
ENST00000549002.1
|
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr17_+_65374075 | 0.28 |
ENST00000581322.1
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr12_-_95611149 | 0.28 |
ENST00000549499.1
ENST00000343958.4 ENST00000546711.1 |
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr11_-_8892464 | 0.28 |
ENST00000527347.1
ENST00000526241.1 ENST00000526126.1 ENST00000530938.1 ENST00000526057.1 |
ST5
|
suppression of tumorigenicity 5 |
| chr11_-_46848393 | 0.25 |
ENST00000526496.1
|
CKAP5
|
cytoskeleton associated protein 5 |
| chr15_+_52311398 | 0.24 |
ENST00000261845.5
|
MAPK6
|
mitogen-activated protein kinase 6 |
| chr4_+_146403912 | 0.24 |
ENST00000507367.1
ENST00000394092.2 ENST00000515385.1 |
SMAD1
|
SMAD family member 1 |
| chr3_-_167813132 | 0.23 |
ENST00000309027.4
|
GOLIM4
|
golgi integral membrane protein 4 |
| chr8_+_79578282 | 0.23 |
ENST00000263849.4
|
ZC2HC1A
|
zinc finger, C2HC-type containing 1A |
| chr6_-_86353510 | 0.22 |
ENST00000444272.1
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr16_+_53242350 | 0.22 |
ENST00000565442.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr12_+_95611569 | 0.22 |
ENST00000261219.6
ENST00000551472.1 ENST00000552821.1 |
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr1_-_26233423 | 0.22 |
ENST00000357865.2
|
STMN1
|
stathmin 1 |
| chr2_-_37899323 | 0.22 |
ENST00000295324.3
ENST00000457889.1 |
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr1_-_26232951 | 0.22 |
ENST00000426559.2
ENST00000455785.2 |
STMN1
|
stathmin 1 |
| chr12_+_95611516 | 0.21 |
ENST00000436874.1
|
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr12_-_104531785 | 0.21 |
ENST00000551727.1
|
NFYB
|
nuclear transcription factor Y, beta |
| chr2_-_160472052 | 0.21 |
ENST00000437839.1
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chrX_+_71401570 | 0.21 |
ENST00000496835.2
ENST00000446576.1 |
PIN4
|
protein (peptidylprolyl cis/trans isomerase) NIMA-interacting, 4 (parvulin) |
| chr19_+_1452188 | 0.20 |
ENST00000587149.1
|
APC2
|
adenomatosis polyposis coli 2 |
| chr1_-_154155675 | 0.20 |
ENST00000330188.9
ENST00000341485.5 |
TPM3
|
tropomyosin 3 |
| chr1_-_91487013 | 0.20 |
ENST00000347275.5
ENST00000370440.1 |
ZNF644
|
zinc finger protein 644 |
| chr9_+_91003271 | 0.20 |
ENST00000375859.3
ENST00000541629.1 |
SPIN1
|
spindlin 1 |
| chrX_+_150866779 | 0.20 |
ENST00000370353.3
|
PRRG3
|
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane) |
| chr4_-_83351294 | 0.20 |
ENST00000502762.1
|
HNRNPDL
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr15_+_77224045 | 0.19 |
ENST00000320963.5
ENST00000394883.3 |
RCN2
|
reticulocalbin 2, EF-hand calcium binding domain |
| chr2_+_219283815 | 0.19 |
ENST00000248444.5
ENST00000454069.1 ENST00000392114.2 |
VIL1
|
villin 1 |
| chr20_+_60698180 | 0.18 |
ENST00000361670.3
|
LSM14B
|
LSM14B, SCD6 homolog B (S. cerevisiae) |
| chr2_-_239149300 | 0.18 |
ENST00000436051.1
|
HES6
|
hes family bHLH transcription factor 6 |
| chr1_+_244998918 | 0.18 |
ENST00000366528.3
|
COX20
|
COX20 cytochrome C oxidase assembly factor |
| chr2_+_149402989 | 0.16 |
ENST00000397424.2
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr5_-_16936340 | 0.16 |
ENST00000507288.1
ENST00000513610.1 |
MYO10
|
myosin X |
| chr10_-_33247124 | 0.16 |
ENST00000414670.1
ENST00000302278.3 ENST00000374956.4 ENST00000488494.1 ENST00000417122.2 ENST00000474568.1 |
ITGB1
|
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) |
| chr6_+_122720681 | 0.16 |
ENST00000368455.4
ENST00000452194.1 |
HSF2
|
heat shock transcription factor 2 |
| chr1_-_154155595 | 0.16 |
ENST00000328159.4
ENST00000368531.2 ENST00000323144.7 ENST00000368533.3 ENST00000341372.3 |
TPM3
|
tropomyosin 3 |
| chr4_+_154387480 | 0.16 |
ENST00000409663.3
ENST00000440693.1 ENST00000409959.3 |
KIAA0922
|
KIAA0922 |
| chr14_-_91884115 | 0.16 |
ENST00000389857.6
|
CCDC88C
|
coiled-coil domain containing 88C |
| chr9_-_74979420 | 0.16 |
ENST00000343431.2
ENST00000376956.3 |
ZFAND5
|
zinc finger, AN1-type domain 5 |
| chr20_+_33292507 | 0.16 |
ENST00000414082.1
|
TP53INP2
|
tumor protein p53 inducible nuclear protein 2 |
| chr4_-_80994210 | 0.16 |
ENST00000403729.2
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr1_-_26232522 | 0.16 |
ENST00000399728.1
|
STMN1
|
stathmin 1 |
| chr12_+_95612006 | 0.15 |
ENST00000551311.1
ENST00000546445.1 |
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr17_-_37764128 | 0.15 |
ENST00000302584.4
|
NEUROD2
|
neuronal differentiation 2 |
| chr19_-_10341948 | 0.15 |
ENST00000590320.1
ENST00000592342.1 ENST00000588952.1 |
S1PR2
DNMT1
|
sphingosine-1-phosphate receptor 2 DNA (cytosine-5-)-methyltransferase 1 |
| chr5_-_114515734 | 0.15 |
ENST00000514154.1
ENST00000282369.3 |
TRIM36
|
tripartite motif containing 36 |
| chr12_-_122985494 | 0.15 |
ENST00000336229.4
|
ZCCHC8
|
zinc finger, CCHC domain containing 8 |
| chr10_+_18948311 | 0.15 |
ENST00000377275.3
|
ARL5B
|
ADP-ribosylation factor-like 5B |
| chr1_+_40915725 | 0.15 |
ENST00000484445.1
ENST00000411995.2 ENST00000361584.3 |
ZFP69B
|
ZFP69 zinc finger protein B |
| chr3_+_52719936 | 0.15 |
ENST00000418458.1
ENST00000394799.2 |
GNL3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr1_+_25071848 | 0.15 |
ENST00000374379.4
|
CLIC4
|
chloride intracellular channel 4 |
| chr1_+_200993071 | 0.15 |
ENST00000446333.1
ENST00000458003.1 |
RP11-168O16.1
|
RP11-168O16.1 |
| chr11_-_95523500 | 0.15 |
ENST00000540054.1
|
FAM76B
|
family with sequence similarity 76, member B |
| chr3_-_167813672 | 0.15 |
ENST00000470487.1
|
GOLIM4
|
golgi integral membrane protein 4 |
| chr6_-_41040195 | 0.15 |
ENST00000463088.1
ENST00000469104.1 ENST00000486443.1 |
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chrX_+_70503526 | 0.14 |
ENST00000413858.1
ENST00000450092.1 |
NONO
|
non-POU domain containing, octamer-binding |
| chr3_+_141105235 | 0.14 |
ENST00000503809.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr19_+_49467232 | 0.14 |
ENST00000599784.1
ENST00000594305.1 |
CTD-2639E6.9
|
CTD-2639E6.9 |
| chr3_+_49977894 | 0.14 |
ENST00000433811.1
|
RBM6
|
RNA binding motif protein 6 |
| chr1_+_68150744 | 0.14 |
ENST00000370986.4
ENST00000370985.3 |
GADD45A
|
growth arrest and DNA-damage-inducible, alpha |
| chr6_+_80714332 | 0.14 |
ENST00000502580.1
ENST00000511260.1 |
TTK
|
TTK protein kinase |
| chr7_-_105925558 | 0.14 |
ENST00000222553.3
|
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chr6_-_41863098 | 0.14 |
ENST00000373006.1
|
USP49
|
ubiquitin specific peptidase 49 |
| chr6_-_27440460 | 0.14 |
ENST00000377419.1
|
ZNF184
|
zinc finger protein 184 |
| chr1_-_95391315 | 0.14 |
ENST00000545882.1
ENST00000415017.1 |
CNN3
|
calponin 3, acidic |
| chr4_-_157892055 | 0.14 |
ENST00000422544.2
|
PDGFC
|
platelet derived growth factor C |
| chr4_-_83350580 | 0.14 |
ENST00000349655.4
ENST00000602300.1 |
HNRNPDL
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr12_-_122985067 | 0.14 |
ENST00000540586.1
ENST00000543897.1 |
ZCCHC8
|
zinc finger, CCHC domain containing 8 |
| chr10_-_128359074 | 0.13 |
ENST00000544758.1
|
C10orf90
|
chromosome 10 open reading frame 90 |
| chr1_+_150229554 | 0.13 |
ENST00000369111.4
|
CA14
|
carbonic anhydrase XIV |
| chr4_+_156775910 | 0.13 |
ENST00000506072.1
ENST00000507590.1 |
TDO2
|
tryptophan 2,3-dioxygenase |
| chr17_+_65373531 | 0.13 |
ENST00000580974.1
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr2_+_46926326 | 0.13 |
ENST00000394861.2
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr4_+_54243862 | 0.13 |
ENST00000306932.6
|
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr18_+_21718924 | 0.13 |
ENST00000399496.3
|
CABYR
|
calcium binding tyrosine-(Y)-phosphorylation regulated |
| chr4_-_80993717 | 0.13 |
ENST00000307333.7
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr6_-_108278456 | 0.13 |
ENST00000429168.1
|
SEC63
|
SEC63 homolog (S. cerevisiae) |
| chr1_-_226496772 | 0.13 |
ENST00000359525.2
ENST00000460719.1 |
LIN9
|
lin-9 homolog (C. elegans) |
| chr17_-_79849438 | 0.13 |
ENST00000331204.4
ENST00000505490.2 |
ALYREF
|
Aly/REF export factor |
| chr1_-_247171347 | 0.13 |
ENST00000339986.7
ENST00000487338.2 |
ZNF695
|
zinc finger protein 695 |
| chr14_+_58862630 | 0.13 |
ENST00000360945.2
|
TOMM20L
|
translocase of outer mitochondrial membrane 20 homolog (yeast)-like |
| chr2_+_61293021 | 0.13 |
ENST00000402291.1
|
KIAA1841
|
KIAA1841 |
| chr12_-_96793142 | 0.13 |
ENST00000552262.1
ENST00000551816.1 ENST00000552496.1 |
CDK17
|
cyclin-dependent kinase 17 |
| chr1_-_93645818 | 0.13 |
ENST00000370280.1
ENST00000479918.1 |
TMED5
|
transmembrane emp24 protein transport domain containing 5 |
| chrX_+_71401526 | 0.13 |
ENST00000218432.5
ENST00000423432.2 ENST00000373669.2 |
PIN4
|
protein (peptidylprolyl cis/trans isomerase) NIMA-interacting, 4 (parvulin) |
| chr18_-_74728998 | 0.13 |
ENST00000359645.3
ENST00000397875.3 ENST00000397869.3 ENST00000578193.1 ENST00000578873.1 ENST00000397866.4 ENST00000528160.1 ENST00000527041.1 ENST00000526111.1 ENST00000397865.5 ENST00000382582.3 |
MBP
|
myelin basic protein |
| chr8_+_104384616 | 0.12 |
ENST00000520337.1
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr4_+_113152881 | 0.12 |
ENST00000274000.5
|
AP1AR
|
adaptor-related protein complex 1 associated regulatory protein |
| chr22_+_31199037 | 0.12 |
ENST00000424224.1
|
OSBP2
|
oxysterol binding protein 2 |
| chr18_+_20494078 | 0.12 |
ENST00000579124.1
ENST00000577588.1 ENST00000582354.1 ENST00000581819.1 |
RBBP8
|
retinoblastoma binding protein 8 |
| chr11_+_110001723 | 0.12 |
ENST00000528673.1
|
ZC3H12C
|
zinc finger CCCH-type containing 12C |
| chrX_+_100646190 | 0.12 |
ENST00000471855.1
|
RPL36A
|
ribosomal protein L36a |
| chr9_+_103235365 | 0.12 |
ENST00000374879.4
|
TMEFF1
|
transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr13_-_41593425 | 0.12 |
ENST00000239882.3
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chr3_+_49977440 | 0.12 |
ENST00000442092.1
ENST00000266022.4 ENST00000443081.1 |
RBM6
|
RNA binding motif protein 6 |
| chr11_-_85430088 | 0.12 |
ENST00000533057.1
ENST00000533892.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr11_-_102323740 | 0.12 |
ENST00000398136.2
|
TMEM123
|
transmembrane protein 123 |
| chr10_-_32217717 | 0.12 |
ENST00000396144.4
ENST00000375245.4 ENST00000344936.2 ENST00000375250.5 |
ARHGAP12
|
Rho GTPase activating protein 12 |
| chr5_-_65017921 | 0.12 |
ENST00000381007.4
|
SGTB
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr9_-_88896977 | 0.12 |
ENST00000311534.6
|
ISCA1
|
iron-sulfur cluster assembly 1 |
| chr4_+_95128996 | 0.12 |
ENST00000457823.2
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr17_-_27277615 | 0.12 |
ENST00000583747.1
ENST00000584236.1 |
PHF12
|
PHD finger protein 12 |
| chr16_-_31085033 | 0.12 |
ENST00000414399.1
|
ZNF668
|
zinc finger protein 668 |
| chr4_+_113152978 | 0.12 |
ENST00000309703.6
|
AP1AR
|
adaptor-related protein complex 1 associated regulatory protein |
| chr4_+_24661479 | 0.11 |
ENST00000569621.1
|
RP11-496D24.2
|
RP11-496D24.2 |
| chr6_-_27440837 | 0.11 |
ENST00000211936.6
|
ZNF184
|
zinc finger protein 184 |
| chr18_-_45456693 | 0.11 |
ENST00000587421.1
|
SMAD2
|
SMAD family member 2 |
| chr14_-_38036271 | 0.11 |
ENST00000556024.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr16_-_75529273 | 0.11 |
ENST00000390664.2
|
CHST6
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 6 |
| chr2_-_55276320 | 0.11 |
ENST00000357376.3
|
RTN4
|
reticulon 4 |
| chr10_-_14596140 | 0.11 |
ENST00000496330.1
|
FAM107B
|
family with sequence similarity 107, member B |
| chr4_+_95129061 | 0.11 |
ENST00000354268.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr14_-_71107921 | 0.11 |
ENST00000553982.1
ENST00000500016.1 |
CTD-2540L5.5
CTD-2540L5.6
|
CTD-2540L5.5 CTD-2540L5.6 |
| chr1_+_93645314 | 0.11 |
ENST00000343253.7
|
CCDC18
|
coiled-coil domain containing 18 |
| chr3_-_79068138 | 0.11 |
ENST00000495273.1
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr4_-_80993854 | 0.11 |
ENST00000346652.6
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr11_+_57365150 | 0.11 |
ENST00000457869.1
ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr4_-_83351005 | 0.11 |
ENST00000295470.5
|
HNRNPDL
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr6_-_41040268 | 0.11 |
ENST00000373154.2
ENST00000244558.9 ENST00000464633.1 ENST00000424266.2 ENST00000479950.1 ENST00000482515.1 |
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chrX_-_135962876 | 0.11 |
ENST00000431446.3
ENST00000570135.1 ENST00000320676.7 ENST00000562646.1 |
RBMX
|
RNA binding motif protein, X-linked |
| chr11_-_102323489 | 0.11 |
ENST00000361236.3
|
TMEM123
|
transmembrane protein 123 |
| chr8_-_80993010 | 0.11 |
ENST00000537855.1
ENST00000520527.1 ENST00000517427.1 ENST00000448733.2 ENST00000379097.3 |
TPD52
|
tumor protein D52 |
| chr2_+_149402553 | 0.11 |
ENST00000258484.6
ENST00000409654.1 |
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chrX_+_80457442 | 0.11 |
ENST00000373212.5
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like |
| chr1_-_225840747 | 0.11 |
ENST00000366843.2
ENST00000366844.3 |
ENAH
|
enabled homolog (Drosophila) |
| chr1_+_145727681 | 0.11 |
ENST00000417171.1
ENST00000451928.2 |
PDZK1
|
PDZ domain containing 1 |
| chr13_+_36920569 | 0.11 |
ENST00000379848.2
|
SPG20OS
|
SPG20 opposite strand |
| chr6_-_134861089 | 0.11 |
ENST00000606039.1
|
RP11-557H15.4
|
RP11-557H15.4 |
| chr15_+_79166065 | 0.11 |
ENST00000559690.1
ENST00000559158.1 |
MORF4L1
|
mortality factor 4 like 1 |
| chr17_-_56065484 | 0.11 |
ENST00000581208.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr17_-_73149921 | 0.11 |
ENST00000481647.1
ENST00000470924.1 |
HN1
|
hematological and neurological expressed 1 |
| chr3_-_52002194 | 0.11 |
ENST00000466412.1
|
PCBP4
|
poly(rC) binding protein 4 |
| chr11_-_95522907 | 0.11 |
ENST00000358780.5
ENST00000542135.1 |
FAM76B
|
family with sequence similarity 76, member B |
| chr16_+_3254247 | 0.11 |
ENST00000304646.2
|
OR1F1
|
olfactory receptor, family 1, subfamily F, member 1 |
| chrX_+_100645812 | 0.11 |
ENST00000427805.2
ENST00000553110.3 ENST00000392994.3 ENST00000409338.1 ENST00000409170.3 |
RPL36A
RPL36A-HNRNPH2
|
ribosomal protein L36a RPL36A-HNRNPH2 readthrough |
| chr4_-_52883786 | 0.10 |
ENST00000343457.3
|
LRRC66
|
leucine rich repeat containing 66 |
| chr10_-_14646388 | 0.10 |
ENST00000468747.1
ENST00000378467.4 |
FAM107B
|
family with sequence similarity 107, member B |
| chr5_-_39425222 | 0.10 |
ENST00000320816.6
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr3_-_71114066 | 0.10 |
ENST00000485326.2
|
FOXP1
|
forkhead box P1 |
| chr11_-_8795787 | 0.10 |
ENST00000528196.1
ENST00000533681.1 |
ST5
|
suppression of tumorigenicity 5 |
| chr10_-_61122220 | 0.10 |
ENST00000422313.2
ENST00000435852.2 ENST00000442566.3 ENST00000373868.2 ENST00000277705.6 ENST00000373867.3 ENST00000419214.2 |
FAM13C
|
family with sequence similarity 13, member C |
| chr18_+_21719018 | 0.10 |
ENST00000585037.1
ENST00000415309.2 ENST00000399481.2 ENST00000577705.1 ENST00000327201.6 |
CABYR
|
calcium binding tyrosine-(Y)-phosphorylation regulated |
| chr14_+_77882741 | 0.10 |
ENST00000595520.1
|
FKSG61
|
FKSG61 |
| chr14_-_91884150 | 0.10 |
ENST00000553403.1
|
CCDC88C
|
coiled-coil domain containing 88C |
| chr7_+_69064566 | 0.10 |
ENST00000403018.2
|
AUTS2
|
autism susceptibility candidate 2 |
| chr5_-_176900610 | 0.10 |
ENST00000477391.2
ENST00000393565.1 ENST00000309007.5 |
DBN1
|
drebrin 1 |
| chr6_+_64282447 | 0.10 |
ENST00000370650.2
ENST00000578299.1 |
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr16_-_46864955 | 0.10 |
ENST00000565112.1
|
C16orf87
|
chromosome 16 open reading frame 87 |
| chr13_+_76334795 | 0.10 |
ENST00000526202.1
ENST00000465261.2 |
LMO7
|
LIM domain 7 |
| chr11_-_6677018 | 0.10 |
ENST00000299441.3
|
DCHS1
|
dachsous cadherin-related 1 |
| chr19_+_3224700 | 0.10 |
ENST00000292672.2
ENST00000541430.2 |
CELF5
|
CUGBP, Elav-like family member 5 |
| chr7_+_16793160 | 0.10 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13 |
| chr11_+_125365110 | 0.10 |
ENST00000527818.1
|
AP000708.1
|
AP000708.1 |
| chr12_+_69979446 | 0.10 |
ENST00000543146.2
|
CCT2
|
chaperonin containing TCP1, subunit 2 (beta) |
| chr3_+_49977523 | 0.10 |
ENST00000422955.1
|
RBM6
|
RNA binding motif protein 6 |
| chr11_+_114271367 | 0.10 |
ENST00000544582.1
ENST00000545678.1 |
RBM7
|
RNA binding motif protein 7 |
| chr9_-_115249484 | 0.10 |
ENST00000457681.1
|
C9orf147
|
chromosome 9 open reading frame 147 |
| chr5_-_114631958 | 0.10 |
ENST00000395557.4
|
CCDC112
|
coiled-coil domain containing 112 |
| chr1_+_228337553 | 0.10 |
ENST00000366714.2
|
GJC2
|
gap junction protein, gamma 2, 47kDa |
| chr3_-_57233966 | 0.10 |
ENST00000473921.1
ENST00000295934.3 |
HESX1
|
HESX homeobox 1 |
| chr6_+_56911476 | 0.10 |
ENST00000545356.1
|
KIAA1586
|
KIAA1586 |
| chr4_+_26323764 | 0.10 |
ENST00000514730.1
ENST00000507574.1 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr19_-_48673465 | 0.10 |
ENST00000598938.1
|
LIG1
|
ligase I, DNA, ATP-dependent |
| chr11_-_85430204 | 0.10 |
ENST00000389958.3
ENST00000527794.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr7_+_7222233 | 0.10 |
ENST00000436587.2
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr3_+_164924716 | 0.10 |
ENST00000470138.1
ENST00000498616.1 |
RP11-85M11.2
|
RP11-85M11.2 |
| chr5_+_179135246 | 0.10 |
ENST00000508787.1
|
CANX
|
calnexin |
| chr21_+_30671690 | 0.10 |
ENST00000399921.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr14_-_57960456 | 0.10 |
ENST00000534126.1
ENST00000422976.2 |
C14orf105
|
chromosome 14 open reading frame 105 |
| chr2_+_170590321 | 0.10 |
ENST00000392647.2
|
KLHL23
|
kelch-like family member 23 |
| chr15_+_57210961 | 0.10 |
ENST00000557843.1
|
TCF12
|
transcription factor 12 |
| chr15_-_50647274 | 0.10 |
ENST00000543881.1
|
GABPB1
|
GA binding protein transcription factor, beta subunit 1 |
| chr12_-_45270151 | 0.10 |
ENST00000429094.2
|
NELL2
|
NEL-like 2 (chicken) |
| chr19_-_37019136 | 0.10 |
ENST00000592282.1
|
ZNF260
|
zinc finger protein 260 |
| chr8_+_63161491 | 0.10 |
ENST00000523211.1
ENST00000524201.1 |
NKAIN3
|
Na+/K+ transporting ATPase interacting 3 |
| chr1_+_212797789 | 0.10 |
ENST00000294829.3
|
FAM71A
|
family with sequence similarity 71, member A |
| chrX_+_135579238 | 0.09 |
ENST00000535601.1
ENST00000448450.1 ENST00000425695.1 |
HTATSF1
|
HIV-1 Tat specific factor 1 |
| chr19_+_55795493 | 0.09 |
ENST00000309383.1
|
BRSK1
|
BR serine/threonine kinase 1 |
| chr2_-_232328867 | 0.09 |
ENST00000453992.1
ENST00000417652.1 ENST00000454824.1 |
NCL
|
nucleolin |
| chrX_-_77395186 | 0.09 |
ENST00000341864.5
|
TAF9B
|
TAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31kDa |
| chr16_+_53241854 | 0.09 |
ENST00000565803.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr1_+_144339738 | 0.09 |
ENST00000538264.1
|
AL592284.1
|
Protein LOC642441 |
| chr12_-_104531945 | 0.09 |
ENST00000551446.1
|
NFYB
|
nuclear transcription factor Y, beta |
| chr10_+_180643 | 0.09 |
ENST00000509513.2
ENST00000397959.3 |
ZMYND11
|
zinc finger, MYND-type containing 11 |
| chr7_+_77167343 | 0.09 |
ENST00000433369.2
ENST00000415482.2 |
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr19_-_18385221 | 0.09 |
ENST00000595654.2
ENST00000593659.1 ENST00000599528.1 |
KIAA1683
|
KIAA1683 |
| chr11_+_34643600 | 0.09 |
ENST00000530286.1
ENST00000533754.1 |
EHF
|
ets homologous factor |
| chr15_+_63335899 | 0.09 |
ENST00000561266.1
|
TPM1
|
tropomyosin 1 (alpha) |
| chr17_-_49198216 | 0.09 |
ENST00000262013.7
ENST00000357122.4 |
SPAG9
|
sperm associated antigen 9 |
| chr4_+_75023816 | 0.09 |
ENST00000395759.2
ENST00000331145.6 ENST00000359107.5 ENST00000325278.6 |
MTHFD2L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2-like |
| chr10_+_70480963 | 0.09 |
ENST00000265872.6
ENST00000535016.1 ENST00000538031.1 ENST00000543719.1 ENST00000539539.1 ENST00000543225.1 ENST00000536012.1 ENST00000494903.2 |
CCAR1
|
cell division cycle and apoptosis regulator 1 |
| chr3_+_49977490 | 0.09 |
ENST00000539992.1
|
RBM6
|
RNA binding motif protein 6 |
| chr12_-_64784306 | 0.09 |
ENST00000543259.1
|
C12orf56
|
chromosome 12 open reading frame 56 |
| chr3_-_149688971 | 0.09 |
ENST00000498307.1
ENST00000489155.1 |
PFN2
|
profilin 2 |
| chr2_+_69240511 | 0.09 |
ENST00000409349.3
|
ANTXR1
|
anthrax toxin receptor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX3_SOX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.1 | 0.3 | GO:0051086 | chaperone mediated protein folding independent of cofactor(GO:0051086) |
| 0.1 | 0.2 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.1 | 0.2 | GO:0036333 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.1 | 0.2 | GO:0033385 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.1 | 0.4 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.1 | 0.2 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.1 | 0.4 | GO:1901297 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.1 | GO:0044771 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.1 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.1 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.0 | 0.3 | GO:0010710 | regulation of collagen catabolic process(GO:0010710) |
| 0.0 | 0.1 | GO:1903461 | Okazaki fragment processing involved in mitotic DNA replication(GO:1903461) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.2 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.0 | GO:1904869 | regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 0.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 | 0.1 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.0 | 0.1 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.0 | 0.1 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.0 | 0.0 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.1 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.0 | 0.1 | GO:0035188 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.2 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.3 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 0.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.2 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.1 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.0 | 0.1 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 | 0.2 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.0 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.0 | 0.1 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.0 | 0.0 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.1 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.3 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0051808 | translocation of peptides or proteins into host(GO:0042000) translocation of peptides or proteins into host cell cytoplasm(GO:0044053) translocation of molecules into host(GO:0044417) translocation of peptides or proteins into other organism involved in symbiotic interaction(GO:0051808) translocation of molecules into other organism involved in symbiotic interaction(GO:0051836) |
| 0.0 | 0.0 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.1 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.0 | 0.1 | GO:0038162 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.0 | 0.1 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) regulation of amacrine cell differentiation(GO:1902869) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.0 | 0.3 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.1 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.1 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 | 0.2 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.0 | 0.1 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 | 0.1 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) |
| 0.0 | 0.0 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.2 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.0 | 0.1 | GO:1903751 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.0 | 0.3 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.1 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.0 | 0.1 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.1 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.0 | 0.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.1 | GO:0090156 | cellular sphingolipid homeostasis(GO:0090156) |
| 0.0 | 0.1 | GO:0043387 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.0 | 0.1 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.1 | GO:1904530 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.0 | 0.0 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.0 | 0.1 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.1 | GO:1990834 | response to odorant(GO:1990834) |
| 0.0 | 0.0 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.0 | 0.1 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 | 0.1 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.1 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.1 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.1 | GO:2000845 | testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.0 | 0.0 | GO:2000584 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.0 | 0.1 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.0 | 0.1 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.0 | GO:0021503 | neural fold bending(GO:0021503) |
| 0.0 | 0.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.0 | 0.0 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.3 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.1 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.1 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.0 | 0.1 | GO:2001162 | regulation of histone H3-K79 methylation(GO:2001160) positive regulation of histone H3-K79 methylation(GO:2001162) |
| 0.0 | 0.1 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.0 | 0.1 | GO:0002415 | immune response in mucosal-associated lymphoid tissue(GO:0002386) immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.0 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.0 | 0.4 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.1 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.2 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) T-helper 1 cell activation(GO:0035711) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.0 | 0.2 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.0 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.2 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.0 | GO:0035284 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.3 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 0.0 | GO:0035750 | protein localization to myelin sheath abaxonal region(GO:0035750) |
| 0.0 | 0.0 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.0 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:0039507 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.0 | 0.2 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.1 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.0 | 0.1 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.0 | 0.0 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.0 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.0 | 0.0 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.0 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.0 | GO:0019520 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.0 | 0.1 | GO:0030865 | cortical cytoskeleton organization(GO:0030865) cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.0 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.0 | 0.0 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.0 | 0.2 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.0 | GO:1901526 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.0 | 0.7 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.2 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.0 | GO:2000395 | positive regulation of viral budding via host ESCRT complex(GO:1903774) regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.0 | GO:1900244 | positive regulation of synaptic vesicle endocytosis(GO:1900244) positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:1901317 | regulation of sperm motility(GO:1901317) |
| 0.0 | 0.1 | GO:0071352 | cellular response to interleukin-2(GO:0071352) |
| 0.0 | 0.1 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.0 | 0.2 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:0061084 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.0 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.1 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.0 | 0.0 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.0 | 0.1 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.2 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.0 | GO:1901340 | negative regulation of store-operated calcium channel activity(GO:1901340) |
| 0.0 | 0.0 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.0 | GO:0070898 | RNA polymerase III transcriptional preinitiation complex assembly(GO:0070898) |
| 0.0 | 0.1 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.2 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.0 | GO:1904300 | positive regulation of neutrophil degranulation(GO:0043315) cellular response to gravity(GO:0071258) positive regulation of neutrophil activation(GO:1902565) regulation of transcytosis(GO:1904298) positive regulation of transcytosis(GO:1904300) regulation of maternal process involved in parturition(GO:1904301) positive regulation of maternal process involved in parturition(GO:1904303) response to 2-O-acetyl-1-O-hexadecyl-sn-glycero-3-phosphocholine(GO:1904316) cellular response to 2-O-acetyl-1-O-hexadecyl-sn-glycero-3-phosphocholine(GO:1904317) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0034677 | integrin alpha7-beta1 complex(GO:0034677) |
| 0.0 | 0.4 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.4 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.6 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.3 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0008623 | CHRAC(GO:0008623) |
| 0.0 | 0.4 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.1 | GO:1990907 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.2 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.1 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.0 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.0 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.0 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.0 | 0.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.1 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.0 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 0.0 | GO:0097637 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.0 | 0.1 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.0 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.0 | 0.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 1.5 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.0 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.0 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.0 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.0 | 0.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.0 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 0.1 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.2 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.2 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.0 | 0.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.3 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.1 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.2 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 0.1 | GO:0017089 | glycolipid transporter activity(GO:0017089) ceramide binding(GO:0097001) |
| 0.0 | 0.1 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.0 | 0.2 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.0 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.1 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.1 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 0.0 | 0.4 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.1 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.0 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.1 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.0 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.1 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.0 | 0.0 | GO:0045142 | triplex DNA binding(GO:0045142) |
| 0.0 | 0.1 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.0 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 0.0 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.2 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.0 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.4 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.0 | GO:0004310 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.1 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.0 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.1 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.0 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.0 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.1 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.0 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.0 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.0 | 0.0 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.6 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.0 | PID S1P S1P4 PATHWAY | S1P4 pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.4 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |