Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
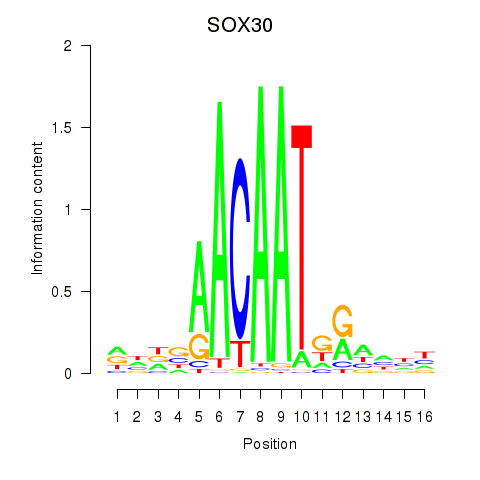
Results for SOX30
Z-value: 0.55
Transcription factors associated with SOX30
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX30
|
ENSG00000039600.6 | SRY-box transcription factor 30 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX30 | hg19_v2_chr5_-_157079372_157079395 | 0.68 | 3.2e-01 | Click! |
Activity profile of SOX30 motif
Sorted Z-values of SOX30 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_11002063 | 0.24 |
ENST00000544994.1
ENST00000228811.4 ENST00000540107.1 |
PRR4
|
proline rich 4 (lacrimal) |
| chr22_+_31488433 | 0.19 |
ENST00000455608.1
|
SMTN
|
smoothelin |
| chr14_+_52456327 | 0.18 |
ENST00000556760.1
|
C14orf166
|
chromosome 14 open reading frame 166 |
| chr11_-_85430088 | 0.15 |
ENST00000533057.1
ENST00000533892.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr14_+_52456193 | 0.14 |
ENST00000261700.3
|
C14orf166
|
chromosome 14 open reading frame 166 |
| chr1_+_244998918 | 0.13 |
ENST00000366528.3
|
COX20
|
COX20 cytochrome C oxidase assembly factor |
| chr1_-_198906528 | 0.12 |
ENST00000432296.1
|
MIR181A1HG
|
MIR181A1 host gene (non-protein coding) |
| chr11_-_102323740 | 0.12 |
ENST00000398136.2
|
TMEM123
|
transmembrane protein 123 |
| chr2_-_162931052 | 0.12 |
ENST00000360534.3
|
DPP4
|
dipeptidyl-peptidase 4 |
| chr1_-_243418621 | 0.12 |
ENST00000366544.1
ENST00000366543.1 |
CEP170
|
centrosomal protein 170kDa |
| chr10_+_70480963 | 0.12 |
ENST00000265872.6
ENST00000535016.1 ENST00000538031.1 ENST00000543719.1 ENST00000539539.1 ENST00000543225.1 ENST00000536012.1 ENST00000494903.2 |
CCAR1
|
cell division cycle and apoptosis regulator 1 |
| chr1_-_243418344 | 0.12 |
ENST00000366542.1
|
CEP170
|
centrosomal protein 170kDa |
| chr1_+_228337553 | 0.12 |
ENST00000366714.2
|
GJC2
|
gap junction protein, gamma 2, 47kDa |
| chr11_-_102323489 | 0.12 |
ENST00000361236.3
|
TMEM123
|
transmembrane protein 123 |
| chr11_-_85430204 | 0.11 |
ENST00000389958.3
ENST00000527794.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr11_-_85430163 | 0.11 |
ENST00000529581.1
ENST00000533577.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr2_-_37899323 | 0.09 |
ENST00000295324.3
ENST00000457889.1 |
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr1_+_185014496 | 0.09 |
ENST00000367510.3
|
RNF2
|
ring finger protein 2 |
| chr12_-_49581152 | 0.09 |
ENST00000550811.1
|
TUBA1A
|
tubulin, alpha 1a |
| chr5_+_179135246 | 0.09 |
ENST00000508787.1
|
CANX
|
calnexin |
| chr10_+_35416223 | 0.09 |
ENST00000489321.1
ENST00000427847.2 ENST00000345491.3 ENST00000395895.2 ENST00000374728.3 ENST00000487132.1 |
CREM
|
cAMP responsive element modulator |
| chr1_-_26231589 | 0.09 |
ENST00000374291.1
|
STMN1
|
stathmin 1 |
| chr4_+_54243798 | 0.09 |
ENST00000337488.6
ENST00000358575.5 ENST00000507922.1 |
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr4_+_113152978 | 0.09 |
ENST00000309703.6
|
AP1AR
|
adaptor-related protein complex 1 associated regulatory protein |
| chr5_+_67588312 | 0.08 |
ENST00000519025.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr4_+_54243917 | 0.08 |
ENST00000507166.1
|
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr17_-_38821373 | 0.08 |
ENST00000394052.3
|
KRT222
|
keratin 222 |
| chr4_+_54243862 | 0.07 |
ENST00000306932.6
|
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr3_-_165555200 | 0.07 |
ENST00000479451.1
ENST00000540653.1 ENST00000488954.1 ENST00000264381.3 |
BCHE
|
butyrylcholinesterase |
| chr5_+_139175380 | 0.07 |
ENST00000274710.3
|
PSD2
|
pleckstrin and Sec7 domain containing 2 |
| chr12_+_95611536 | 0.07 |
ENST00000549002.1
|
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr4_-_69111401 | 0.07 |
ENST00000332644.5
|
TMPRSS11B
|
transmembrane protease, serine 11B |
| chr14_-_36645674 | 0.07 |
ENST00000556013.2
|
PTCSC3
|
papillary thyroid carcinoma susceptibility candidate 3 (non-protein coding) |
| chr6_-_128222103 | 0.06 |
ENST00000434358.1
ENST00000543064.1 ENST00000368248.2 |
THEMIS
|
thymocyte selection associated |
| chr11_+_125365110 | 0.06 |
ENST00000527818.1
|
AP000708.1
|
AP000708.1 |
| chrX_+_102192200 | 0.06 |
ENST00000218249.5
|
RAB40AL
|
RAB40A, member RAS oncogene family-like |
| chr10_+_35416090 | 0.06 |
ENST00000354759.3
|
CREM
|
cAMP responsive element modulator |
| chr12_+_95611516 | 0.06 |
ENST00000436874.1
|
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr9_-_134145880 | 0.06 |
ENST00000372269.3
ENST00000464831.1 |
FAM78A
|
family with sequence similarity 78, member A |
| chr1_-_243418650 | 0.06 |
ENST00000522995.1
|
CEP170
|
centrosomal protein 170kDa |
| chr12_+_95611569 | 0.06 |
ENST00000261219.6
ENST00000551472.1 ENST00000552821.1 |
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr10_+_35415851 | 0.06 |
ENST00000374726.3
|
CREM
|
cAMP responsive element modulator |
| chr13_+_98795664 | 0.06 |
ENST00000376581.5
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr11_-_18548426 | 0.06 |
ENST00000357193.3
ENST00000536719.1 |
TSG101
|
tumor susceptibility 101 |
| chrX_-_15872914 | 0.05 |
ENST00000380291.1
ENST00000545766.1 ENST00000421527.2 ENST00000329235.2 |
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr21_+_35552978 | 0.05 |
ENST00000428914.2
ENST00000609062.1 ENST00000609947.1 |
LINC00310
|
long intergenic non-protein coding RNA 310 |
| chr17_-_73149921 | 0.05 |
ENST00000481647.1
ENST00000470924.1 |
HN1
|
hematological and neurological expressed 1 |
| chr20_-_5426380 | 0.05 |
ENST00000609252.1
ENST00000422352.1 |
LINC00658
|
long intergenic non-protein coding RNA 658 |
| chr17_+_47448102 | 0.05 |
ENST00000576461.1
|
RP11-81K2.1
|
Uncharacterized protein |
| chr9_-_85882145 | 0.05 |
ENST00000328788.1
|
FRMD3
|
FERM domain containing 3 |
| chr3_-_19988462 | 0.05 |
ENST00000344838.4
|
EFHB
|
EF-hand domain family, member B |
| chr16_+_4545853 | 0.05 |
ENST00000575129.1
ENST00000398595.3 ENST00000414777.1 |
HMOX2
|
heme oxygenase (decycling) 2 |
| chr13_-_41593425 | 0.05 |
ENST00000239882.3
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chr2_+_99953816 | 0.05 |
ENST00000289371.6
|
EIF5B
|
eukaryotic translation initiation factor 5B |
| chr5_-_146461027 | 0.05 |
ENST00000394410.2
ENST00000508267.1 ENST00000504198.1 |
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr21_+_30671690 | 0.04 |
ENST00000399921.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr8_-_141810634 | 0.04 |
ENST00000521986.1
ENST00000523539.1 ENST00000538769.1 |
PTK2
|
protein tyrosine kinase 2 |
| chr20_-_62610982 | 0.04 |
ENST00000369886.3
ENST00000450107.1 |
SAMD10
|
sterile alpha motif domain containing 10 |
| chr5_+_65222438 | 0.04 |
ENST00000380938.2
|
ERBB2IP
|
erbb2 interacting protein |
| chr1_-_21503337 | 0.04 |
ENST00000400422.1
ENST00000602326.1 ENST00000411888.1 ENST00000438975.1 |
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr18_+_7946941 | 0.04 |
ENST00000444013.1
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M |
| chr2_-_169769787 | 0.04 |
ENST00000451987.1
|
SPC25
|
SPC25, NDC80 kinetochore complex component |
| chr1_+_40942887 | 0.04 |
ENST00000372706.1
|
ZFP69
|
ZFP69 zinc finger protein |
| chr2_+_27719697 | 0.04 |
ENST00000264717.2
ENST00000424318.2 |
GCKR
|
glucokinase (hexokinase 4) regulator |
| chr7_-_82792215 | 0.04 |
ENST00000333891.9
ENST00000423517.2 |
PCLO
|
piccolo presynaptic cytomatrix protein |
| chrY_+_15016013 | 0.04 |
ENST00000360160.4
ENST00000454054.1 |
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr3_-_122283424 | 0.04 |
ENST00000477522.2
ENST00000360356.2 |
PARP9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr9_+_103235365 | 0.04 |
ENST00000374879.4
|
TMEFF1
|
transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr15_+_93426514 | 0.04 |
ENST00000556722.1
|
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr22_+_46449674 | 0.04 |
ENST00000381051.2
|
FLJ27365
|
hsa-mir-4763 |
| chr6_-_11779014 | 0.04 |
ENST00000229583.5
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr20_-_45981138 | 0.04 |
ENST00000446994.2
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr2_-_157198860 | 0.03 |
ENST00000409572.1
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr17_+_67498538 | 0.03 |
ENST00000589647.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr11_-_85430356 | 0.03 |
ENST00000526999.1
|
SYTL2
|
synaptotagmin-like 2 |
| chr6_-_11779174 | 0.03 |
ENST00000379413.2
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr14_+_36295504 | 0.03 |
ENST00000216807.7
|
BRMS1L
|
breast cancer metastasis-suppressor 1-like |
| chr2_+_86669118 | 0.03 |
ENST00000427678.1
ENST00000542128.1 |
KDM3A
|
lysine (K)-specific demethylase 3A |
| chr1_+_93913713 | 0.03 |
ENST00000604705.1
ENST00000370253.2 |
FNBP1L
|
formin binding protein 1-like |
| chr1_+_244998602 | 0.03 |
ENST00000411948.2
|
COX20
|
COX20 cytochrome C oxidase assembly factor |
| chr1_-_200379129 | 0.03 |
ENST00000367353.1
|
ZNF281
|
zinc finger protein 281 |
| chr5_+_170288856 | 0.03 |
ENST00000523189.1
|
RANBP17
|
RAN binding protein 17 |
| chr1_+_149239529 | 0.03 |
ENST00000457216.2
|
RP11-403I13.4
|
RP11-403I13.4 |
| chr6_+_12290586 | 0.03 |
ENST00000379375.5
|
EDN1
|
endothelin 1 |
| chr20_-_5426332 | 0.03 |
ENST00000420529.1
|
LINC00658
|
long intergenic non-protein coding RNA 658 |
| chr12_-_99288986 | 0.03 |
ENST00000552407.1
ENST00000551613.1 ENST00000548447.1 ENST00000546364.3 ENST00000552748.1 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr9_-_74383302 | 0.03 |
ENST00000377066.5
|
TMEM2
|
transmembrane protein 2 |
| chr21_+_35553045 | 0.03 |
ENST00000416145.1
ENST00000430922.1 ENST00000419881.2 |
LINC00310
|
long intergenic non-protein coding RNA 310 |
| chr5_+_135468516 | 0.03 |
ENST00000507118.1
ENST00000511116.1 ENST00000545279.1 ENST00000545620.1 |
SMAD5
|
SMAD family member 5 |
| chr15_+_57211318 | 0.03 |
ENST00000557947.1
|
TCF12
|
transcription factor 12 |
| chr1_+_47489240 | 0.03 |
ENST00000371901.3
|
CYP4X1
|
cytochrome P450, family 4, subfamily X, polypeptide 1 |
| chr11_-_89224638 | 0.02 |
ENST00000535633.1
ENST00000263317.4 |
NOX4
|
NADPH oxidase 4 |
| chr4_+_144303093 | 0.02 |
ENST00000505913.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr5_+_38148582 | 0.02 |
ENST00000508853.1
|
CTD-2207A17.1
|
CTD-2207A17.1 |
| chr11_-_8964580 | 0.02 |
ENST00000325884.1
|
ASCL3
|
achaete-scute family bHLH transcription factor 3 |
| chr5_-_141338627 | 0.02 |
ENST00000231484.3
|
PCDH12
|
protocadherin 12 |
| chr22_-_43485381 | 0.02 |
ENST00000331018.7
ENST00000266254.7 ENST00000445824.1 |
TTLL1
|
tubulin tyrosine ligase-like family, member 1 |
| chr2_+_143635067 | 0.02 |
ENST00000264170.4
|
KYNU
|
kynureninase |
| chr6_+_126240442 | 0.02 |
ENST00000448104.1
ENST00000438495.1 ENST00000444128.1 |
NCOA7
|
nuclear receptor coactivator 7 |
| chr6_+_22146851 | 0.02 |
ENST00000606197.1
|
CASC15
|
cancer susceptibility candidate 15 (non-protein coding) |
| chrX_+_114874727 | 0.02 |
ENST00000543070.1
|
PLS3
|
plastin 3 |
| chr3_-_8686479 | 0.02 |
ENST00000544814.1
ENST00000427408.1 |
SSUH2
|
ssu-2 homolog (C. elegans) |
| chrX_+_41192595 | 0.02 |
ENST00000399959.2
|
DDX3X
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked |
| chr3_+_19988566 | 0.02 |
ENST00000273047.4
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr18_-_53804580 | 0.01 |
ENST00000590484.1
ENST00000589293.1 ENST00000587904.1 ENST00000591974.1 |
RP11-456O19.4
|
RP11-456O19.4 |
| chr1_-_167883353 | 0.01 |
ENST00000545172.1
|
ADCY10
|
adenylate cyclase 10 (soluble) |
| chr2_-_166060382 | 0.01 |
ENST00000409101.3
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr5_-_59481406 | 0.01 |
ENST00000546160.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr6_-_134639180 | 0.01 |
ENST00000367858.5
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr22_-_30970498 | 0.01 |
ENST00000431313.1
|
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr4_+_187148556 | 0.01 |
ENST00000264690.6
ENST00000446598.2 ENST00000414291.1 ENST00000513864.1 |
KLKB1
|
kallikrein B, plasma (Fletcher factor) 1 |
| chr2_+_232573208 | 0.01 |
ENST00000409115.3
|
PTMA
|
prothymosin, alpha |
| chr1_-_156828810 | 0.00 |
ENST00000368195.3
|
INSRR
|
insulin receptor-related receptor |
| chr12_+_27623565 | 0.00 |
ENST00000535986.1
|
SMCO2
|
single-pass membrane protein with coiled-coil domains 2 |
| chr12_+_9066472 | 0.00 |
ENST00000538657.1
|
PHC1
|
polyhomeotic homolog 1 (Drosophila) |
| chr21_+_30672433 | 0.00 |
ENST00000451655.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr4_-_87028478 | 0.00 |
ENST00000515400.1
ENST00000395157.3 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr1_+_185703513 | 0.00 |
ENST00000271588.4
ENST00000367492.2 |
HMCN1
|
hemicentin 1 |
| chr12_+_80750134 | 0.00 |
ENST00000546620.1
|
OTOGL
|
otogelin-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX30
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.0 | 0.4 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.1 | GO:0036343 | negative regulation of extracellular matrix disassembly(GO:0010716) psychomotor behavior(GO:0036343) |
| 0.0 | 0.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:2000395 | positive regulation of viral budding via host ESCRT complex(GO:1903774) regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.3 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.3 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.0 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 0.0 | GO:0051866 | general adaptation syndrome(GO:0051866) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 0.1 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.0 | 0.0 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.0 | GO:0031705 | bombesin receptor binding(GO:0031705) |