Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
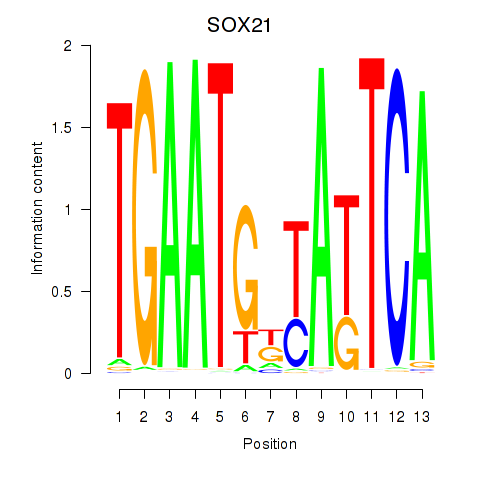
Results for SOX21
Z-value: 0.24
Transcription factors associated with SOX21
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX21
|
ENSG00000125285.4 | SRY-box transcription factor 21 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX21 | hg19_v2_chr13_-_95364389_95364389 | -0.74 | 2.6e-01 | Click! |
Activity profile of SOX21 motif
Sorted Z-values of SOX21 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_42381173 | 0.11 |
ENST00000221972.3
|
CD79A
|
CD79a molecule, immunoglobulin-associated alpha |
| chr3_+_186288454 | 0.11 |
ENST00000265028.3
|
DNAJB11
|
DnaJ (Hsp40) homolog, subfamily B, member 11 |
| chr2_+_3705785 | 0.10 |
ENST00000252505.3
|
ALLC
|
allantoicase |
| chr19_-_2783255 | 0.10 |
ENST00000589251.1
|
SGTA
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha |
| chr11_+_35201826 | 0.09 |
ENST00000531873.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr14_+_64565442 | 0.09 |
ENST00000553308.1
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2 |
| chr19_+_42381337 | 0.08 |
ENST00000597454.1
ENST00000444740.2 |
CD79A
|
CD79a molecule, immunoglobulin-associated alpha |
| chr15_-_80263506 | 0.08 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr16_-_15149828 | 0.08 |
ENST00000566419.1
ENST00000568320.1 |
NTAN1
|
N-terminal asparagine amidase |
| chr10_-_12084770 | 0.07 |
ENST00000357604.5
|
UPF2
|
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr4_-_103747011 | 0.07 |
ENST00000350435.7
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr22_+_39353527 | 0.07 |
ENST00000249116.2
|
APOBEC3A
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3A |
| chr6_+_31371337 | 0.07 |
ENST00000449934.2
ENST00000421350.1 |
MICA
|
MHC class I polypeptide-related sequence A |
| chrY_+_14813160 | 0.06 |
ENST00000338981.3
|
USP9Y
|
ubiquitin specific peptidase 9, Y-linked |
| chr5_+_67588312 | 0.06 |
ENST00000519025.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr5_+_134181755 | 0.06 |
ENST00000504727.1
ENST00000435259.2 ENST00000508791.1 |
C5orf24
|
chromosome 5 open reading frame 24 |
| chr10_-_18944123 | 0.06 |
ENST00000606425.1
|
RP11-139J15.7
|
Uncharacterized protein |
| chr4_-_103746683 | 0.06 |
ENST00000504211.1
ENST00000508476.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr8_+_67687413 | 0.06 |
ENST00000521960.1
ENST00000522398.1 ENST00000522629.1 ENST00000520976.1 ENST00000396596.1 |
SGK3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr17_+_66245341 | 0.06 |
ENST00000577985.1
|
AMZ2
|
archaelysin family metallopeptidase 2 |
| chr14_-_102771462 | 0.06 |
ENST00000522874.1
|
MOK
|
MOK protein kinase |
| chr4_-_103746924 | 0.06 |
ENST00000505207.1
ENST00000502404.1 ENST00000507845.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr14_-_92588013 | 0.06 |
ENST00000553514.1
ENST00000605997.1 |
NDUFB1
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 1, 7kDa |
| chr8_+_26150628 | 0.05 |
ENST00000523925.1
ENST00000315985.7 |
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha |
| chr4_-_83931862 | 0.05 |
ENST00000506560.1
ENST00000442461.2 ENST00000446851.2 ENST00000340417.3 |
LIN54
|
lin-54 homolog (C. elegans) |
| chr4_+_75174204 | 0.05 |
ENST00000332112.4
ENST00000514968.1 ENST00000503098.1 ENST00000502358.1 ENST00000509145.1 ENST00000505212.1 |
EPGN
|
epithelial mitogen |
| chr3_-_45957534 | 0.05 |
ENST00000536047.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr1_+_203764742 | 0.05 |
ENST00000432282.1
ENST00000453771.1 ENST00000367214.1 ENST00000367212.3 ENST00000332127.4 |
ZC3H11A
|
zinc finger CCCH-type containing 11A |
| chr8_+_101170563 | 0.05 |
ENST00000520508.1
ENST00000388798.2 |
SPAG1
|
sperm associated antigen 1 |
| chr12_-_89746173 | 0.05 |
ENST00000308385.6
|
DUSP6
|
dual specificity phosphatase 6 |
| chr1_+_152881014 | 0.05 |
ENST00000368764.3
ENST00000392667.2 |
IVL
|
involucrin |
| chr7_-_37382683 | 0.04 |
ENST00000455879.1
|
ELMO1
|
engulfment and cell motility 1 |
| chr14_-_38028689 | 0.04 |
ENST00000553425.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr3_-_164875850 | 0.04 |
ENST00000472120.1
|
RP11-747D18.1
|
RP11-747D18.1 |
| chr4_-_39034542 | 0.04 |
ENST00000344606.6
|
TMEM156
|
transmembrane protein 156 |
| chr3_-_37034702 | 0.04 |
ENST00000322716.5
|
EPM2AIP1
|
EPM2A (laforin) interacting protein 1 |
| chr8_-_143961236 | 0.04 |
ENST00000377675.3
ENST00000517471.1 ENST00000292427.4 |
CYP11B1
|
cytochrome P450, family 11, subfamily B, polypeptide 1 |
| chr6_+_151815143 | 0.04 |
ENST00000239374.7
ENST00000367290.5 |
CCDC170
|
coiled-coil domain containing 170 |
| chr3_-_45957088 | 0.04 |
ENST00000539217.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr19_-_2783363 | 0.03 |
ENST00000221566.2
|
SGTA
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha |
| chr6_-_121655593 | 0.03 |
ENST00000398212.2
|
TBC1D32
|
TBC1 domain family, member 32 |
| chr1_-_202776392 | 0.03 |
ENST00000235790.4
|
KDM5B
|
lysine (K)-specific demethylase 5B |
| chr16_-_15982440 | 0.03 |
ENST00000575938.1
ENST00000573396.1 ENST00000573968.1 ENST00000575744.1 ENST00000573429.1 ENST00000255759.6 ENST00000575073.1 |
FOPNL
|
FGFR1OP N-terminal like |
| chr1_-_169455169 | 0.03 |
ENST00000367804.4
ENST00000236137.5 |
SLC19A2
|
solute carrier family 19 (thiamine transporter), member 2 |
| chr22_-_40929812 | 0.03 |
ENST00000422851.1
|
MKL1
|
megakaryoblastic leukemia (translocation) 1 |
| chr14_+_92588281 | 0.03 |
ENST00000298875.4
ENST00000553427.1 |
CPSF2
|
cleavage and polyadenylation specific factor 2, 100kDa |
| chr8_-_94029882 | 0.03 |
ENST00000520686.1
|
TRIQK
|
triple QxxK/R motif containing |
| chr12_+_101673872 | 0.02 |
ENST00000261637.4
|
UTP20
|
UTP20, small subunit (SSU) processome component, homolog (yeast) |
| chr12_+_123011776 | 0.02 |
ENST00000450485.2
ENST00000333479.7 |
KNTC1
|
kinetochore associated 1 |
| chr1_-_54303934 | 0.02 |
ENST00000537333.1
|
NDC1
|
NDC1 transmembrane nucleoporin |
| chr14_-_92588246 | 0.02 |
ENST00000329559.3
|
NDUFB1
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 1, 7kDa |
| chr14_+_50291993 | 0.02 |
ENST00000595378.1
|
AL627171.2
|
HCG1786899; PRO2610; Uncharacterized protein |
| chr6_-_121655850 | 0.02 |
ENST00000422369.1
|
TBC1D32
|
TBC1 domain family, member 32 |
| chr11_+_2482661 | 0.02 |
ENST00000335475.5
|
KCNQ1
|
potassium voltage-gated channel, KQT-like subfamily, member 1 |
| chr3_+_141103634 | 0.02 |
ENST00000507722.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr1_+_146714291 | 0.02 |
ENST00000431239.1
ENST00000369259.3 ENST00000369258.4 ENST00000361293.5 |
CHD1L
|
chromodomain helicase DNA binding protein 1-like |
| chr3_-_141747459 | 0.02 |
ENST00000477292.1
ENST00000478006.1 ENST00000495310.1 ENST00000486111.1 |
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr6_-_154567984 | 0.02 |
ENST00000517438.1
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1 |
| chr7_-_14942944 | 0.02 |
ENST00000403951.2
|
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr11_+_114549108 | 0.02 |
ENST00000389586.4
ENST00000375475.5 |
NXPE2
|
neurexophilin and PC-esterase domain family, member 2 |
| chr3_-_141747439 | 0.01 |
ENST00000467667.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr14_+_60712463 | 0.01 |
ENST00000325642.3
ENST00000529574.1 |
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr1_+_151171012 | 0.01 |
ENST00000349792.5
ENST00000409426.1 ENST00000441902.2 ENST00000368890.4 ENST00000424999.1 ENST00000368888.4 |
PIP5K1A
|
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha |
| chr6_+_42531798 | 0.01 |
ENST00000372903.2
ENST00000372899.1 ENST00000372901.1 |
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr18_+_29769978 | 0.01 |
ENST00000269202.6
ENST00000581447.1 |
MEP1B
|
meprin A, beta |
| chr3_-_155572164 | 0.01 |
ENST00000392845.3
ENST00000359479.3 |
SLC33A1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr10_-_17243579 | 0.01 |
ENST00000525762.1
ENST00000412821.3 ENST00000351358.4 ENST00000377766.5 ENST00000358282.7 ENST00000488990.1 ENST00000377799.3 |
TRDMT1
|
tRNA aspartic acid methyltransferase 1 |
| chr3_+_37035289 | 0.01 |
ENST00000455445.2
ENST00000441265.1 ENST00000435176.1 ENST00000429117.1 ENST00000536378.1 |
MLH1
|
mutL homolog 1 |
| chr5_+_175875349 | 0.01 |
ENST00000261942.6
|
FAF2
|
Fas associated factor family member 2 |
| chr12_-_1058849 | 0.01 |
ENST00000358495.3
|
RAD52
|
RAD52 homolog (S. cerevisiae) |
| chr18_-_47018897 | 0.01 |
ENST00000418495.1
|
RPL17
|
ribosomal protein L17 |
| chr1_-_151148492 | 0.01 |
ENST00000295314.4
|
TMOD4
|
tropomodulin 4 (muscle) |
| chrX_-_55208866 | 0.01 |
ENST00000545075.1
|
MTRNR2L10
|
MT-RNR2-like 10 |
| chr10_-_71169031 | 0.01 |
ENST00000373307.1
|
TACR2
|
tachykinin receptor 2 |
| chr5_-_179045199 | 0.01 |
ENST00000523921.1
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr19_+_18682531 | 0.01 |
ENST00000596304.1
ENST00000430157.2 |
UBA52
|
ubiquitin A-52 residue ribosomal protein fusion product 1 |
| chr7_+_2394445 | 0.01 |
ENST00000360876.4
ENST00000413917.1 ENST00000397011.2 |
EIF3B
|
eukaryotic translation initiation factor 3, subunit B |
| chr1_-_75100539 | 0.01 |
ENST00000420661.2
|
C1orf173
|
chromosome 1 open reading frame 173 |
| chrX_-_131353461 | 0.01 |
ENST00000370874.1
|
RAP2C
|
RAP2C, member of RAS oncogene family |
| chr6_-_52926539 | 0.00 |
ENST00000350082.5
ENST00000356971.3 |
ICK
|
intestinal cell (MAK-like) kinase |
| chr4_-_69083720 | 0.00 |
ENST00000432593.3
|
TMPRSS11BNL
|
TMPRSS11B N-terminal like |
| chr1_+_158901329 | 0.00 |
ENST00000368140.1
ENST00000368138.3 ENST00000392254.2 ENST00000392252.3 ENST00000368135.4 |
PYHIN1
|
pyrin and HIN domain family, member 1 |
| chr3_+_37035263 | 0.00 |
ENST00000458205.2
ENST00000539477.1 |
MLH1
|
mutL homolog 1 |
| chrY_+_16634483 | 0.00 |
ENST00000382872.1
|
NLGN4Y
|
neuroligin 4, Y-linked |
| chr4_-_177190364 | 0.00 |
ENST00000296525.3
|
ASB5
|
ankyrin repeat and SOCS box containing 5 |
| chr12_-_1058685 | 0.00 |
ENST00000397230.2
ENST00000542785.1 ENST00000544742.1 ENST00000536177.1 ENST00000539046.1 ENST00000541619.1 |
RAD52
|
RAD52 homolog (S. cerevisiae) |
| chr12_-_10324716 | 0.00 |
ENST00000545927.1
ENST00000432556.2 ENST00000309539.3 ENST00000544577.1 |
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr21_+_30672433 | 0.00 |
ENST00000451655.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr5_-_95158644 | 0.00 |
ENST00000237858.6
|
GLRX
|
glutaredoxin (thioltransferase) |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX21
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 0.0 | 0.1 | GO:1903568 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.1 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 0.0 | GO:2000861 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 0.0 | 0.1 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.1 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |