Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
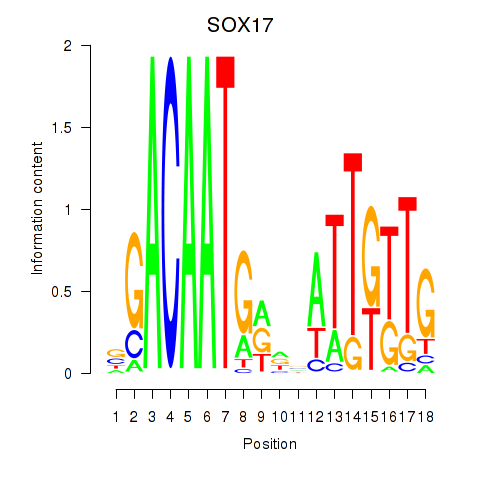
Results for SOX17
Z-value: 1.37
Transcription factors associated with SOX17
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX17
|
ENSG00000164736.5 | SRY-box transcription factor 17 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX17 | hg19_v2_chr8_+_55370487_55370503 | -0.61 | 3.9e-01 | Click! |
Activity profile of SOX17 motif
Sorted Z-values of SOX17 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_104029362 | 0.61 |
ENST00000495778.1
|
APOPT1
|
apoptogenic 1, mitochondrial |
| chr14_+_77882741 | 0.58 |
ENST00000595520.1
|
FKSG61
|
FKSG61 |
| chr22_-_32766972 | 0.55 |
ENST00000382084.4
ENST00000382086.2 |
RFPL3S
|
RFPL3 antisense |
| chr9_+_86237963 | 0.54 |
ENST00000277124.8
|
IDNK
|
idnK, gluconokinase homolog (E. coli) |
| chr19_+_58514229 | 0.51 |
ENST00000546949.1
ENST00000553254.1 ENST00000547364.1 |
CTD-2368P22.1
|
HCG1811579; Uncharacterized protein |
| chr5_+_81601166 | 0.50 |
ENST00000439350.1
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like |
| chr12_+_52668394 | 0.47 |
ENST00000423955.2
|
KRT86
|
keratin 86 |
| chr14_-_36645674 | 0.43 |
ENST00000556013.2
|
PTCSC3
|
papillary thyroid carcinoma susceptibility candidate 3 (non-protein coding) |
| chr4_-_74864386 | 0.41 |
ENST00000296027.4
|
CXCL5
|
chemokine (C-X-C motif) ligand 5 |
| chr12_+_7053228 | 0.41 |
ENST00000540506.2
|
C12orf57
|
chromosome 12 open reading frame 57 |
| chr11_+_94300474 | 0.40 |
ENST00000299001.6
|
PIWIL4
|
piwi-like RNA-mediated gene silencing 4 |
| chr4_-_15939963 | 0.37 |
ENST00000259988.2
|
FGFBP1
|
fibroblast growth factor binding protein 1 |
| chr4_+_103790462 | 0.36 |
ENST00000503643.1
|
CISD2
|
CDGSH iron sulfur domain 2 |
| chr2_-_157186630 | 0.36 |
ENST00000406048.2
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr14_+_101361107 | 0.36 |
ENST00000553584.1
ENST00000554852.1 |
MEG8
|
maternally expressed 8 (non-protein coding) |
| chr18_-_24283586 | 0.35 |
ENST00000579458.1
|
U3
|
Small nucleolar RNA U3 |
| chr1_+_227751231 | 0.35 |
ENST00000343776.5
ENST00000608949.1 ENST00000397097.3 |
ZNF678
|
zinc finger protein 678 |
| chr21_+_29911640 | 0.35 |
ENST00000412526.1
ENST00000455939.1 |
LINC00161
|
long intergenic non-protein coding RNA 161 |
| chr7_+_112120908 | 0.34 |
ENST00000439068.2
ENST00000312849.4 ENST00000429049.1 |
LSMEM1
|
leucine-rich single-pass membrane protein 1 |
| chrX_-_55187588 | 0.34 |
ENST00000472571.2
ENST00000332132.4 ENST00000425133.2 ENST00000358460.4 |
FAM104B
|
family with sequence similarity 104, member B |
| chr17_+_57297807 | 0.34 |
ENST00000284116.4
ENST00000581140.1 ENST00000581276.1 |
GDPD1
|
glycerophosphodiester phosphodiesterase domain containing 1 |
| chr2_-_219906220 | 0.34 |
ENST00000458526.1
ENST00000409865.3 ENST00000410037.1 ENST00000457968.1 ENST00000436631.1 ENST00000341552.5 ENST00000441968.1 ENST00000295729.2 |
CCDC108
|
coiled-coil domain containing 108 |
| chr3_-_45837959 | 0.34 |
ENST00000353278.4
ENST00000456124.2 |
SLC6A20
|
solute carrier family 6 (proline IMINO transporter), member 20 |
| chr3_-_45838011 | 0.34 |
ENST00000358525.4
ENST00000413781.1 |
SLC6A20
|
solute carrier family 6 (proline IMINO transporter), member 20 |
| chr2_-_220034745 | 0.32 |
ENST00000455516.2
ENST00000396775.3 ENST00000295738.7 |
SLC23A3
|
solute carrier family 23, member 3 |
| chr2_+_71357744 | 0.32 |
ENST00000498451.2
|
MPHOSPH10
|
M-phase phosphoprotein 10 (U3 small nucleolar ribonucleoprotein) |
| chr2_+_234637754 | 0.32 |
ENST00000482026.1
ENST00000609767.1 |
UGT1A3
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A3 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr11_+_29181503 | 0.31 |
ENST00000530960.1
|
RP11-466I1.1
|
RP11-466I1.1 |
| chr6_-_26216872 | 0.30 |
ENST00000244601.3
|
HIST1H2BG
|
histone cluster 1, H2bg |
| chr20_-_44207965 | 0.30 |
ENST00000357199.4
ENST00000289953.2 |
WFDC8
|
WAP four-disulfide core domain 8 |
| chr11_+_32605350 | 0.30 |
ENST00000531120.1
ENST00000524896.1 ENST00000323213.5 |
EIF3M
|
eukaryotic translation initiation factor 3, subunit M |
| chr5_+_49962772 | 0.29 |
ENST00000281631.5
ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr11_+_65266507 | 0.29 |
ENST00000544868.1
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr14_+_104029278 | 0.28 |
ENST00000472726.2
ENST00000409074.2 ENST00000440963.1 ENST00000556253.2 ENST00000247618.4 |
RP11-73M18.2
APOPT1
|
Kinesin light chain 1 apoptogenic 1, mitochondrial |
| chr2_+_64073187 | 0.28 |
ENST00000491621.1
|
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr5_+_8839844 | 0.28 |
ENST00000510229.1
ENST00000506655.1 ENST00000510067.1 |
RP11-143A12.3
|
RP11-143A12.3 |
| chr22_+_44464923 | 0.28 |
ENST00000404989.1
|
PARVB
|
parvin, beta |
| chr10_-_37891859 | 0.27 |
ENST00000544824.1
|
MTRNR2L7
|
MT-RNR2-like 7 |
| chr13_+_28519343 | 0.26 |
ENST00000381026.3
|
ATP5EP2
|
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit pseudogene 2 |
| chr3_+_98451275 | 0.26 |
ENST00000265261.6
ENST00000497008.1 |
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr5_+_138678131 | 0.26 |
ENST00000394795.2
ENST00000510080.1 |
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr20_+_47662805 | 0.25 |
ENST00000262982.2
ENST00000542325.1 |
CSE1L
|
CSE1 chromosome segregation 1-like (yeast) |
| chr12_-_40499661 | 0.25 |
ENST00000280871.4
|
SLC2A13
|
solute carrier family 2 (facilitated glucose transporter), member 13 |
| chr11_-_125365435 | 0.25 |
ENST00000524435.1
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr11_-_7695437 | 0.25 |
ENST00000533558.1
ENST00000527542.1 ENST00000531096.1 |
CYB5R2
|
cytochrome b5 reductase 2 |
| chr2_+_71357434 | 0.25 |
ENST00000244230.2
|
MPHOSPH10
|
M-phase phosphoprotein 10 (U3 small nucleolar ribonucleoprotein) |
| chr12_+_7053172 | 0.25 |
ENST00000229281.5
|
C12orf57
|
chromosome 12 open reading frame 57 |
| chr16_-_30023615 | 0.25 |
ENST00000564979.1
ENST00000563378.1 |
DOC2A
|
double C2-like domains, alpha |
| chrX_-_55187531 | 0.24 |
ENST00000489298.1
ENST00000477847.2 |
FAM104B
|
family with sequence similarity 104, member B |
| chr2_+_201677390 | 0.24 |
ENST00000447069.1
|
BZW1
|
basic leucine zipper and W2 domains 1 |
| chr12_-_76461249 | 0.24 |
ENST00000551524.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr19_-_49956728 | 0.24 |
ENST00000601825.1
ENST00000596049.1 ENST00000599366.1 ENST00000597415.1 |
PIH1D1
|
PIH1 domain containing 1 |
| chr3_-_52488048 | 0.24 |
ENST00000232975.3
|
TNNC1
|
troponin C type 1 (slow) |
| chr12_-_111358372 | 0.24 |
ENST00000548438.1
ENST00000228841.8 |
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow |
| chr22_+_24951436 | 0.24 |
ENST00000215829.3
|
SNRPD3
|
small nuclear ribonucleoprotein D3 polypeptide 18kDa |
| chr7_-_99277610 | 0.23 |
ENST00000343703.5
ENST00000222982.4 ENST00000439761.1 ENST00000339843.2 |
CYP3A5
|
cytochrome P450, family 3, subfamily A, polypeptide 5 |
| chr1_+_156589051 | 0.23 |
ENST00000255039.1
|
HAPLN2
|
hyaluronan and proteoglycan link protein 2 |
| chr6_+_7590413 | 0.23 |
ENST00000342415.5
|
SNRNP48
|
small nuclear ribonucleoprotein 48kDa (U11/U12) |
| chr19_-_16606988 | 0.23 |
ENST00000269881.3
|
CALR3
|
calreticulin 3 |
| chr15_-_81616446 | 0.22 |
ENST00000302824.6
|
STARD5
|
StAR-related lipid transfer (START) domain containing 5 |
| chr22_+_32754139 | 0.21 |
ENST00000382088.3
|
RFPL3
|
ret finger protein-like 3 |
| chr20_+_34556488 | 0.21 |
ENST00000373973.3
ENST00000349339.1 ENST00000538900.1 |
CNBD2
|
cyclic nucleotide binding domain containing 2 |
| chr3_-_88108192 | 0.21 |
ENST00000309534.6
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr4_-_170897045 | 0.21 |
ENST00000508313.1
|
RP11-205M3.3
|
RP11-205M3.3 |
| chr17_+_57784826 | 0.21 |
ENST00000262291.4
|
VMP1
|
vacuole membrane protein 1 |
| chr12_+_9822331 | 0.20 |
ENST00000545918.1
ENST00000543300.1 ENST00000261339.6 ENST00000466035.2 |
CLEC2D
|
C-type lectin domain family 2, member D |
| chrX_+_49216659 | 0.20 |
ENST00000415752.1
|
GAGE12I
|
G antigen 12I |
| chr22_+_32753854 | 0.20 |
ENST00000249007.4
|
RFPL3
|
ret finger protein-like 3 |
| chr1_-_19811132 | 0.20 |
ENST00000433834.1
|
CAPZB
|
capping protein (actin filament) muscle Z-line, beta |
| chr18_+_1099004 | 0.20 |
ENST00000581556.1
|
RP11-78F17.1
|
RP11-78F17.1 |
| chr5_+_667759 | 0.20 |
ENST00000594226.1
|
AC026740.1
|
Uncharacterized protein |
| chr18_+_20513782 | 0.20 |
ENST00000399722.2
ENST00000399725.2 ENST00000399721.2 ENST00000583594.1 |
RBBP8
|
retinoblastoma binding protein 8 |
| chr12_-_110888103 | 0.19 |
ENST00000426440.1
ENST00000228825.7 |
ARPC3
|
actin related protein 2/3 complex, subunit 3, 21kDa |
| chr19_-_55720824 | 0.19 |
ENST00000376350.3
ENST00000263434.5 |
PTPRH
|
protein tyrosine phosphatase, receptor type, H |
| chr15_+_69373210 | 0.19 |
ENST00000435479.1
ENST00000559870.1 |
LINC00277
RP11-809H16.5
|
long intergenic non-protein coding RNA 277 RP11-809H16.5 |
| chr11_-_125366018 | 0.19 |
ENST00000527534.1
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr2_+_219283815 | 0.19 |
ENST00000248444.5
ENST00000454069.1 ENST00000392114.2 |
VIL1
|
villin 1 |
| chr15_+_93749295 | 0.19 |
ENST00000599897.1
|
AC112693.2
|
AC112693.2 |
| chr12_-_125398602 | 0.18 |
ENST00000541272.1
ENST00000535131.1 |
UBC
|
ubiquitin C |
| chr3_-_107099454 | 0.18 |
ENST00000593837.1
ENST00000599431.1 |
RP11-446H18.5
|
RP11-446H18.5 |
| chrY_+_2709906 | 0.18 |
ENST00000430575.1
|
RPS4Y1
|
ribosomal protein S4, Y-linked 1 |
| chr16_+_28770025 | 0.18 |
ENST00000435324.2
|
NPIPB9
|
nuclear pore complex interacting protein family, member B9 |
| chr12_+_128399965 | 0.18 |
ENST00000540882.1
ENST00000542089.1 |
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chr2_-_114205429 | 0.18 |
ENST00000440617.1
|
AC016745.1
|
HDCMB45P; Uncharacterized protein |
| chr9_-_70866572 | 0.18 |
ENST00000417734.1
|
AL353608.1
|
HDCMB45P; Uncharacterized protein |
| chr11_-_102668879 | 0.18 |
ENST00000315274.6
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase) |
| chr15_+_34310428 | 0.17 |
ENST00000557872.1
|
CHRM5
|
cholinergic receptor, muscarinic 5 |
| chr4_-_141348763 | 0.17 |
ENST00000509477.1
|
CLGN
|
calmegin |
| chr10_-_91295304 | 0.17 |
ENST00000341233.4
ENST00000371790.4 |
SLC16A12
|
solute carrier family 16, member 12 |
| chr2_-_27851843 | 0.17 |
ENST00000324364.3
|
CCDC121
|
coiled-coil domain containing 121 |
| chr22_+_42017987 | 0.17 |
ENST00000405506.1
|
XRCC6
|
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr19_-_48753028 | 0.17 |
ENST00000522431.1
|
CARD8
|
caspase recruitment domain family, member 8 |
| chr10_+_81462983 | 0.17 |
ENST00000448135.1
ENST00000429828.1 ENST00000372321.1 |
NUTM2B
|
NUT family member 2B |
| chr12_+_16500037 | 0.17 |
ENST00000536371.1
ENST00000010404.2 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr11_-_118122996 | 0.17 |
ENST00000525386.1
ENST00000527472.1 ENST00000278949.4 |
MPZL3
|
myelin protein zero-like 3 |
| chr9_+_69252845 | 0.16 |
ENST00000354995.3
|
BX255923.1
|
HDCMB45P; Uncharacterized protein |
| chr12_-_96794330 | 0.16 |
ENST00000261211.3
|
CDK17
|
cyclin-dependent kinase 17 |
| chr4_-_170679024 | 0.16 |
ENST00000393381.2
|
C4orf27
|
chromosome 4 open reading frame 27 |
| chr4_+_155665123 | 0.16 |
ENST00000336356.3
|
LRAT
|
lecithin retinol acyltransferase (phosphatidylcholine--retinol O-acyltransferase) |
| chr2_+_86668464 | 0.16 |
ENST00000409064.1
|
KDM3A
|
lysine (K)-specific demethylase 3A |
| chrX_-_40036520 | 0.16 |
ENST00000406200.2
ENST00000378455.4 ENST00000342274.4 |
BCOR
|
BCL6 corepressor |
| chr4_+_146402346 | 0.16 |
ENST00000514778.1
ENST00000507594.1 |
SMAD1
|
SMAD family member 1 |
| chr19_-_13030071 | 0.16 |
ENST00000293695.7
|
SYCE2
|
synaptonemal complex central element protein 2 |
| chr6_+_167704838 | 0.16 |
ENST00000366829.2
|
UNC93A
|
unc-93 homolog A (C. elegans) |
| chr6_+_126277842 | 0.16 |
ENST00000229633.5
|
HINT3
|
histidine triad nucleotide binding protein 3 |
| chr3_-_58200398 | 0.15 |
ENST00000318316.3
ENST00000460422.1 ENST00000483681.1 |
DNASE1L3
|
deoxyribonuclease I-like 3 |
| chr13_+_37581115 | 0.15 |
ENST00000481013.1
|
EXOSC8
|
exosome component 8 |
| chr16_+_33629600 | 0.15 |
ENST00000562905.2
|
IGHV3OR16-13
|
immunoglobulin heavy variable 3/OR16-13 (non-functional) |
| chr19_-_49540073 | 0.15 |
ENST00000301407.7
ENST00000601167.1 ENST00000604577.1 ENST00000591656.1 |
CGB1
CTB-60B18.6
|
chorionic gonadotropin, beta polypeptide 1 Choriogonadotropin subunit beta variant 1; Uncharacterized protein |
| chr19_-_19887185 | 0.15 |
ENST00000590092.1
ENST00000589910.1 ENST00000589508.1 ENST00000586645.1 ENST00000586816.1 ENST00000588223.1 ENST00000586885.1 |
LINC00663
|
long intergenic non-protein coding RNA 663 |
| chr2_-_74735707 | 0.15 |
ENST00000233630.6
|
PCGF1
|
polycomb group ring finger 1 |
| chr16_-_15474904 | 0.15 |
ENST00000534094.1
|
NPIPA5
|
nuclear pore complex interacting protein family, member A5 |
| chr15_-_28778117 | 0.15 |
ENST00000525590.2
ENST00000329523.6 |
GOLGA8G
|
golgin A8 family, member G |
| chr7_-_38948774 | 0.15 |
ENST00000395969.2
ENST00000414632.1 ENST00000310301.4 |
VPS41
|
vacuolar protein sorting 41 homolog (S. cerevisiae) |
| chr2_-_75788428 | 0.15 |
ENST00000432649.1
|
EVA1A
|
eva-1 homolog A (C. elegans) |
| chr2_-_38303218 | 0.15 |
ENST00000407341.1
ENST00000260630.3 |
CYP1B1
|
cytochrome P450, family 1, subfamily B, polypeptide 1 |
| chr15_-_47426320 | 0.15 |
ENST00000557832.1
|
FKSG62
|
FKSG62 |
| chrX_+_49178536 | 0.15 |
ENST00000442437.2
|
GAGE12J
|
G antigen 12J |
| chr7_+_16828866 | 0.14 |
ENST00000597084.1
|
AC073333.1
|
Uncharacterized protein |
| chr5_+_121297650 | 0.14 |
ENST00000339397.4
|
SRFBP1
|
serum response factor binding protein 1 |
| chr12_-_11150474 | 0.14 |
ENST00000538986.1
|
TAS2R20
|
taste receptor, type 2, member 20 |
| chr11_+_17316870 | 0.14 |
ENST00000458064.2
|
NUCB2
|
nucleobindin 2 |
| chr8_-_141810634 | 0.14 |
ENST00000521986.1
ENST00000523539.1 ENST00000538769.1 |
PTK2
|
protein tyrosine kinase 2 |
| chr11_-_114430570 | 0.14 |
ENST00000251921.2
|
NXPE1
|
neurexophilin and PC-esterase domain family, member 1 |
| chr11_-_57417405 | 0.14 |
ENST00000524669.1
ENST00000300022.3 |
YPEL4
|
yippee-like 4 (Drosophila) |
| chr19_-_35454953 | 0.14 |
ENST00000404801.1
|
ZNF792
|
zinc finger protein 792 |
| chr20_+_61448376 | 0.14 |
ENST00000343916.3
|
COL9A3
|
collagen, type IX, alpha 3 |
| chr5_+_44809027 | 0.14 |
ENST00000507110.1
|
MRPS30
|
mitochondrial ribosomal protein S30 |
| chr1_+_59775752 | 0.13 |
ENST00000371212.1
|
FGGY
|
FGGY carbohydrate kinase domain containing |
| chr17_+_11924129 | 0.13 |
ENST00000353533.5
ENST00000415385.3 |
MAP2K4
|
mitogen-activated protein kinase kinase 4 |
| chr17_-_56605341 | 0.13 |
ENST00000583114.1
|
SEPT4
|
septin 4 |
| chr2_+_64068074 | 0.13 |
ENST00000394417.2
ENST00000484142.1 ENST00000482668.1 ENST00000467648.2 |
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr5_+_167913450 | 0.13 |
ENST00000231572.3
ENST00000538719.1 |
RARS
|
arginyl-tRNA synthetase |
| chr2_-_86094764 | 0.13 |
ENST00000393808.3
|
ST3GAL5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr7_+_92158083 | 0.13 |
ENST00000265732.5
ENST00000481551.1 ENST00000496410.1 |
RBM48
|
RNA binding motif protein 48 |
| chr4_+_95128996 | 0.13 |
ENST00000457823.2
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr12_-_102455846 | 0.13 |
ENST00000545679.1
|
CCDC53
|
coiled-coil domain containing 53 |
| chr8_-_17942432 | 0.13 |
ENST00000381733.4
ENST00000314146.10 |
ASAH1
|
N-acylsphingosine amidohydrolase (acid ceramidase) 1 |
| chr2_+_234627424 | 0.13 |
ENST00000373409.3
|
UGT1A4
|
UDP glucuronosyltransferase 1 family, polypeptide A4 |
| chr17_+_20978854 | 0.13 |
ENST00000456235.1
|
AC087393.1
|
AC087393.1 |
| chr17_-_58042075 | 0.12 |
ENST00000305783.8
ENST00000589113.1 ENST00000442346.2 |
RNFT1
|
ring finger protein, transmembrane 1 |
| chr11_+_64794991 | 0.12 |
ENST00000352068.5
ENST00000525648.1 |
SNX15
|
sorting nexin 15 |
| chr3_-_46000146 | 0.12 |
ENST00000438446.1
|
FYCO1
|
FYVE and coiled-coil domain containing 1 |
| chr1_+_174417095 | 0.12 |
ENST00000367685.2
|
GPR52
|
G protein-coupled receptor 52 |
| chr19_-_10764509 | 0.12 |
ENST00000591501.1
|
ILF3-AS1
|
ILF3 antisense RNA 1 (head to head) |
| chr7_+_99006232 | 0.12 |
ENST00000403633.2
|
BUD31
|
BUD31 homolog (S. cerevisiae) |
| chr6_-_45345597 | 0.12 |
ENST00000371460.1
ENST00000371459.1 |
SUPT3H
|
suppressor of Ty 3 homolog (S. cerevisiae) |
| chr8_-_57906362 | 0.12 |
ENST00000262644.4
|
IMPAD1
|
inositol monophosphatase domain containing 1 |
| chr4_+_95129061 | 0.12 |
ENST00000354268.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr3_+_88108381 | 0.12 |
ENST00000473136.1
|
RP11-159G9.5
|
Uncharacterized protein |
| chr2_-_225362533 | 0.12 |
ENST00000451538.1
|
CUL3
|
cullin 3 |
| chr11_-_795286 | 0.12 |
ENST00000533385.1
ENST00000527723.1 |
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr12_+_26164645 | 0.12 |
ENST00000542004.1
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr11_+_107879459 | 0.12 |
ENST00000393094.2
|
CUL5
|
cullin 5 |
| chr3_-_105588231 | 0.12 |
ENST00000545639.1
ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr5_-_43557791 | 0.11 |
ENST00000338972.4
ENST00000511321.1 ENST00000515338.1 |
PAIP1
|
poly(A) binding protein interacting protein 1 |
| chr8_-_66474884 | 0.11 |
ENST00000520902.1
|
CTD-3025N20.2
|
CTD-3025N20.2 |
| chr20_-_33539655 | 0.11 |
ENST00000451957.2
|
GSS
|
glutathione synthetase |
| chr19_+_58095501 | 0.11 |
ENST00000536878.2
ENST00000597850.1 ENST00000597219.1 ENST00000598689.1 ENST00000599456.1 ENST00000307468.4 |
ZIK1
|
zinc finger protein interacting with K protein 1 |
| chr16_+_16484691 | 0.11 |
ENST00000344087.4
|
NPIPA7
|
nuclear pore complex interacting protein family, member A7 |
| chr4_+_2794750 | 0.11 |
ENST00000452765.2
ENST00000389838.2 |
SH3BP2
|
SH3-domain binding protein 2 |
| chr5_-_10308125 | 0.11 |
ENST00000296658.3
|
CMBL
|
carboxymethylenebutenolidase homolog (Pseudomonas) |
| chr1_+_145292806 | 0.11 |
ENST00000448873.2
|
NBPF10
|
neuroblastoma breakpoint family, member 10 |
| chr2_+_233527443 | 0.11 |
ENST00000410095.1
|
EFHD1
|
EF-hand domain family, member D1 |
| chr2_-_220034712 | 0.11 |
ENST00000409370.2
ENST00000430764.1 ENST00000409878.3 |
SLC23A3
|
solute carrier family 23, member 3 |
| chr5_-_1345199 | 0.11 |
ENST00000320895.5
|
CLPTM1L
|
CLPTM1-like |
| chr22_-_32589466 | 0.11 |
ENST00000248980.4
|
RFPL2
|
ret finger protein-like 2 |
| chr14_-_93673353 | 0.11 |
ENST00000556566.1
ENST00000306954.4 |
C14orf142
|
chromosome 14 open reading frame 142 |
| chr17_+_27181956 | 0.11 |
ENST00000254928.5
ENST00000580917.2 |
ERAL1
|
Era-like 12S mitochondrial rRNA chaperone 1 |
| chr5_+_142286887 | 0.11 |
ENST00000451259.1
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr1_-_201476274 | 0.10 |
ENST00000340006.2
|
CSRP1
|
cysteine and glycine-rich protein 1 |
| chr6_-_143771799 | 0.10 |
ENST00000237283.8
|
ADAT2
|
adenosine deaminase, tRNA-specific 2 |
| chr19_-_48752812 | 0.10 |
ENST00000359009.4
|
CARD8
|
caspase recruitment domain family, member 8 |
| chr10_-_99052382 | 0.10 |
ENST00000453547.2
ENST00000316676.8 ENST00000358308.3 ENST00000466484.1 ENST00000358531.4 |
ARHGAP19-SLIT1
ARHGAP19
|
ARHGAP19-SLIT1 readthrough (NMD candidate) Rho GTPase activating protein 19 |
| chr1_-_160492994 | 0.10 |
ENST00000368055.1
ENST00000368057.3 ENST00000368059.3 |
SLAMF6
|
SLAM family member 6 |
| chr1_-_45805607 | 0.10 |
ENST00000372104.1
ENST00000448481.1 ENST00000483127.1 ENST00000528013.2 ENST00000456914.2 |
MUTYH
|
mutY homolog |
| chr12_+_70219052 | 0.10 |
ENST00000552032.2
ENST00000547771.2 |
MYRFL
|
myelin regulatory factor-like |
| chr2_+_27851863 | 0.10 |
ENST00000264718.3
ENST00000610189.1 |
GPN1
|
GPN-loop GTPase 1 |
| chr3_-_64009102 | 0.10 |
ENST00000478185.1
ENST00000482510.1 ENST00000497323.1 ENST00000492933.1 ENST00000295901.4 |
PSMD6
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 |
| chr16_+_22517166 | 0.10 |
ENST00000356156.3
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr1_+_109265033 | 0.10 |
ENST00000445274.1
|
FNDC7
|
fibronectin type III domain containing 7 |
| chr11_+_60145948 | 0.10 |
ENST00000300184.3
ENST00000358246.1 |
MS4A7
|
membrane-spanning 4-domains, subfamily A, member 7 |
| chr12_-_76462713 | 0.10 |
ENST00000552056.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr17_-_30228678 | 0.10 |
ENST00000261708.4
|
UTP6
|
UTP6, small subunit (SSU) processome component, homolog (yeast) |
| chr6_+_142468361 | 0.10 |
ENST00000367630.4
|
VTA1
|
vesicle (multivesicular body) trafficking 1 |
| chr10_-_135379132 | 0.10 |
ENST00000343131.5
|
SYCE1
|
synaptonemal complex central element protein 1 |
| chr1_+_66796401 | 0.10 |
ENST00000528771.1
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chrX_+_49235708 | 0.10 |
ENST00000381725.1
|
GAGE2B
|
G antigen 2B |
| chr8_+_27631903 | 0.09 |
ENST00000305188.8
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr12_+_128399917 | 0.09 |
ENST00000544645.1
|
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chr7_-_107642348 | 0.09 |
ENST00000393561.1
|
LAMB1
|
laminin, beta 1 |
| chr17_-_57784755 | 0.09 |
ENST00000537860.1
ENST00000393038.2 ENST00000409433.2 |
PTRH2
|
peptidyl-tRNA hydrolase 2 |
| chr7_+_5085452 | 0.09 |
ENST00000353796.3
ENST00000396912.1 ENST00000396904.2 |
RBAK
RBAK-RBAKDN
|
RB-associated KRAB zinc finger RBAK-RBAKDN readthrough |
| chr19_-_48753104 | 0.09 |
ENST00000447740.2
|
CARD8
|
caspase recruitment domain family, member 8 |
| chr2_-_157198860 | 0.09 |
ENST00000409572.1
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr6_-_19804973 | 0.09 |
ENST00000457670.1
ENST00000607810.1 ENST00000606628.1 |
RP4-625H18.2
|
RP4-625H18.2 |
| chr5_-_16509101 | 0.09 |
ENST00000399793.2
|
FAM134B
|
family with sequence similarity 134, member B |
| chr10_-_69597915 | 0.09 |
ENST00000225171.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr5_+_112073544 | 0.09 |
ENST00000257430.4
ENST00000508376.2 |
APC
|
adenomatous polyposis coli |
| chr4_+_144312659 | 0.09 |
ENST00000509992.1
|
GAB1
|
GRB2-associated binding protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX17
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.1 | 0.5 | GO:0019521 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.1 | 1.0 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.1 | 0.4 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.1 | 0.1 | GO:0046660 | female gonad development(GO:0008585) development of primary female sexual characteristics(GO:0046545) female sex differentiation(GO:0046660) |
| 0.1 | 0.2 | GO:1903939 | negative regulation of histone H3-K9 dimethylation(GO:1900110) regulation of TORC2 signaling(GO:1903939) |
| 0.1 | 0.2 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.1 | 0.4 | GO:0006789 | bilirubin conjugation(GO:0006789) |
| 0.1 | 0.2 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.1 | 0.7 | GO:0015824 | proline transport(GO:0015824) |
| 0.1 | 0.2 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.2 | GO:0071684 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.2 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.0 | 0.4 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.2 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.0 | 0.2 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.2 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.2 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.1 | GO:0045957 | regulation of complement activation, alternative pathway(GO:0030451) negative regulation of complement activation, alternative pathway(GO:0045957) positive regulation of serine-type endopeptidase activity(GO:1900005) positive regulation of serine-type peptidase activity(GO:1902573) |
| 0.0 | 0.1 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.0 | 0.1 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.4 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.2 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.0 | 0.2 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.0 | 0.2 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.1 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.0 | 0.2 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 | 0.1 | GO:1900276 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.0 | 0.3 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.0 | 0.2 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.4 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.2 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.1 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) Golgi to plasma membrane CFTR protein transport(GO:0043000) |
| 0.0 | 0.2 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.4 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.1 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.3 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.2 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.1 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.1 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.8 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.0 | GO:0016098 | response to ozone(GO:0010193) monoterpenoid metabolic process(GO:0016098) |
| 0.0 | 0.5 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.1 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.4 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0042737 | drug catabolic process(GO:0042737) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 0.4 | GO:0071547 | piP-body(GO:0071547) |
| 0.1 | 0.2 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.2 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.3 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.3 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.5 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0043257 | laminin-8 complex(GO:0043257) |
| 0.0 | 0.2 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 0.1 | GO:0072517 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.0 | 0.2 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.3 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.3 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.1 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.1 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005365 | myo-inositol transmembrane transporter activity(GO:0005365) |
| 0.1 | 0.4 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 0.4 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.1 | 0.3 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.1 | 0.2 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.3 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.0 | 0.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.0 | 0.1 | GO:0004513 | neolactotetraosylceramide alpha-2,3-sialyltransferase activity(GO:0004513) lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.0 | 0.1 | GO:0008441 | 3'(2'),5'-bisphosphate nucleotidase activity(GO:0008441) |
| 0.0 | 0.2 | GO:0032408 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.0 | 0.4 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0004877 | complement component C4b receptor activity(GO:0001861) complement component C3b receptor activity(GO:0004877) |
| 0.0 | 0.7 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.2 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.9 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.2 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.2 | GO:0016416 | O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0033961 | cis-stilbene-oxide hydrolase activity(GO:0033961) |
| 0.0 | 0.1 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 0.1 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.0 | 0.2 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.4 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.2 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.3 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.0 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.1 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.5 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.1 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |