Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
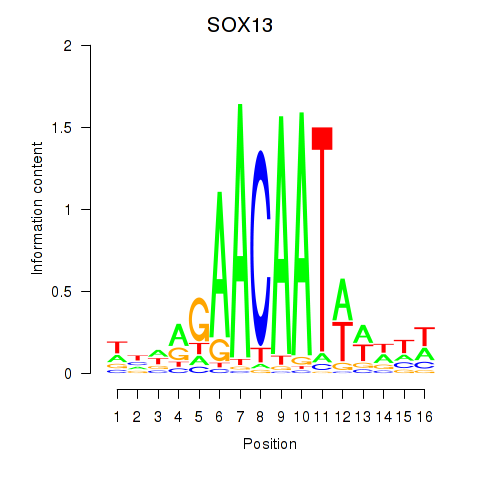
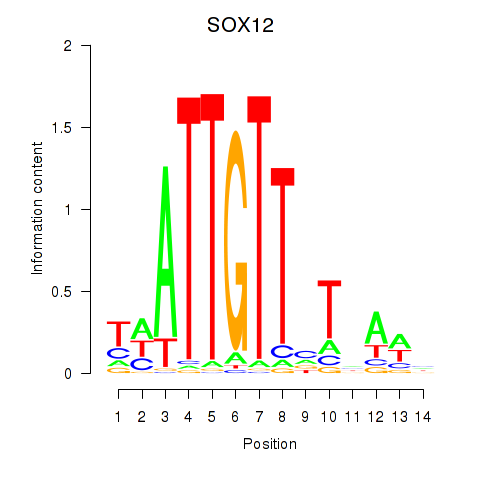
Results for SOX13_SOX12
Z-value: 1.08
Transcription factors associated with SOX13_SOX12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX13
|
ENSG00000143842.10 | SRY-box transcription factor 13 |
|
SOX12
|
ENSG00000177732.6 | SRY-box transcription factor 12 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX12 | hg19_v2_chr20_+_306221_306239 | -0.78 | 2.2e-01 | Click! |
| SOX13 | hg19_v2_chr1_+_204042723_204042784 | 0.49 | 5.1e-01 | Click! |
Activity profile of SOX13_SOX12 motif
Sorted Z-values of SOX13_SOX12 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_134861089 | 0.64 |
ENST00000606039.1
|
RP11-557H15.4
|
RP11-557H15.4 |
| chrX_+_108780062 | 0.53 |
ENST00000372106.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr11_-_85393886 | 0.49 |
ENST00000534224.1
|
CREBZF
|
CREB/ATF bZIP transcription factor |
| chr5_+_136070614 | 0.46 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1 |
| chr8_+_32579271 | 0.46 |
ENST00000518084.1
|
NRG1
|
neuregulin 1 |
| chr7_-_64023441 | 0.45 |
ENST00000309683.6
|
ZNF680
|
zinc finger protein 680 |
| chr12_+_11187087 | 0.44 |
ENST00000601123.1
|
AC018630.1
|
Uncharacterized protein |
| chr19_+_39786962 | 0.44 |
ENST00000333625.2
|
IFNL1
|
interferon, lambda 1 |
| chr8_-_56986768 | 0.40 |
ENST00000523936.1
|
RPS20
|
ribosomal protein S20 |
| chr5_-_114632307 | 0.39 |
ENST00000506442.1
ENST00000379611.5 |
CCDC112
|
coiled-coil domain containing 112 |
| chr20_+_32250079 | 0.39 |
ENST00000375222.3
|
C20orf144
|
chromosome 20 open reading frame 144 |
| chrX_-_77395186 | 0.37 |
ENST00000341864.5
|
TAF9B
|
TAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31kDa |
| chr2_+_179318295 | 0.37 |
ENST00000442710.1
|
DFNB59
|
deafness, autosomal recessive 59 |
| chr10_-_14596140 | 0.36 |
ENST00000496330.1
|
FAM107B
|
family with sequence similarity 107, member B |
| chr12_-_85430024 | 0.34 |
ENST00000547836.1
ENST00000532498.2 |
TSPAN19
|
tetraspanin 19 |
| chr1_-_243418344 | 0.33 |
ENST00000366542.1
|
CEP170
|
centrosomal protein 170kDa |
| chr4_-_74864386 | 0.33 |
ENST00000296027.4
|
CXCL5
|
chemokine (C-X-C motif) ligand 5 |
| chr15_-_79383102 | 0.32 |
ENST00000558480.2
ENST00000419573.3 |
RASGRF1
|
Ras protein-specific guanine nucleotide-releasing factor 1 |
| chr12_-_8803128 | 0.32 |
ENST00000543467.1
|
MFAP5
|
microfibrillar associated protein 5 |
| chr6_-_131299929 | 0.32 |
ENST00000531356.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr1_-_243418621 | 0.32 |
ENST00000366544.1
ENST00000366543.1 |
CEP170
|
centrosomal protein 170kDa |
| chr4_+_54243917 | 0.32 |
ENST00000507166.1
|
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr7_-_112635675 | 0.32 |
ENST00000447785.1
ENST00000451962.1 |
AC018464.3
|
AC018464.3 |
| chr12_-_68845417 | 0.31 |
ENST00000542875.1
|
RP11-81H14.2
|
RP11-81H14.2 |
| chrX_+_108780347 | 0.31 |
ENST00000372103.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr4_+_165675197 | 0.31 |
ENST00000515485.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr1_+_185014496 | 0.30 |
ENST00000367510.3
|
RNF2
|
ring finger protein 2 |
| chr6_+_122720681 | 0.30 |
ENST00000368455.4
ENST00000452194.1 |
HSF2
|
heat shock transcription factor 2 |
| chr4_+_95128996 | 0.30 |
ENST00000457823.2
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr4_+_54243862 | 0.30 |
ENST00000306932.6
|
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr7_-_64023410 | 0.29 |
ENST00000447137.2
|
ZNF680
|
zinc finger protein 680 |
| chr3_+_20081515 | 0.29 |
ENST00000263754.4
|
KAT2B
|
K(lysine) acetyltransferase 2B |
| chr8_+_52730143 | 0.28 |
ENST00000415643.1
|
AC090186.1
|
Uncharacterized protein |
| chr17_+_52978185 | 0.27 |
ENST00000572405.1
ENST00000572158.1 ENST00000540336.1 ENST00000572298.1 ENST00000536554.1 ENST00000575333.1 ENST00000570499.1 ENST00000572576.1 |
TOM1L1
|
target of myb1 (chicken)-like 1 |
| chr11_-_85430088 | 0.27 |
ENST00000533057.1
ENST00000533892.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr4_+_95128748 | 0.27 |
ENST00000359052.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr11_-_85430204 | 0.27 |
ENST00000389958.3
ENST00000527794.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr4_+_78829479 | 0.27 |
ENST00000504901.1
|
MRPL1
|
mitochondrial ribosomal protein L1 |
| chr12_-_49581152 | 0.27 |
ENST00000550811.1
|
TUBA1A
|
tubulin, alpha 1a |
| chr17_+_52978156 | 0.26 |
ENST00000348161.4
|
TOM1L1
|
target of myb1 (chicken)-like 1 |
| chrX_-_115085422 | 0.26 |
ENST00000430756.2
|
RP11-761E20.1
|
RP11-761E20.1 |
| chr4_+_95129061 | 0.26 |
ENST00000354268.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr9_-_86536323 | 0.26 |
ENST00000297814.2
ENST00000413982.1 ENST00000334204.2 |
KIF27
|
kinesin family member 27 |
| chr19_-_23869999 | 0.25 |
ENST00000601935.1
ENST00000359788.4 ENST00000600313.1 ENST00000596211.1 ENST00000599168.1 |
ZNF675
|
zinc finger protein 675 |
| chr5_-_96518907 | 0.25 |
ENST00000508447.1
ENST00000283109.3 |
RIOK2
|
RIO kinase 2 |
| chr1_-_238108575 | 0.25 |
ENST00000604646.1
|
MTRNR2L11
|
MT-RNR2-like 11 (pseudogene) |
| chr18_-_44497308 | 0.25 |
ENST00000585916.1
ENST00000324794.7 ENST00000545673.1 |
PIAS2
|
protein inhibitor of activated STAT, 2 |
| chr5_-_133702761 | 0.25 |
ENST00000521118.1
ENST00000265334.4 ENST00000435211.1 |
CDKL3
|
cyclin-dependent kinase-like 3 |
| chr19_-_57678811 | 0.25 |
ENST00000554048.2
|
DUXA
|
double homeobox A |
| chr4_-_83931862 | 0.25 |
ENST00000506560.1
ENST00000442461.2 ENST00000446851.2 ENST00000340417.3 |
LIN54
|
lin-54 homolog (C. elegans) |
| chr1_+_10057274 | 0.23 |
ENST00000294435.7
|
RBP7
|
retinol binding protein 7, cellular |
| chrX_+_108779004 | 0.23 |
ENST00000218004.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr16_-_15474904 | 0.23 |
ENST00000534094.1
|
NPIPA5
|
nuclear pore complex interacting protein family, member A5 |
| chr12_+_7917812 | 0.23 |
ENST00000382119.1
|
NANOGNB
|
NANOG neighbor homeobox |
| chr2_-_201729284 | 0.22 |
ENST00000434813.2
|
CLK1
|
CDC-like kinase 1 |
| chr16_-_18462221 | 0.22 |
ENST00000528301.1
|
RP11-1212A22.4
|
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr11_+_74303612 | 0.22 |
ENST00000527458.1
ENST00000532497.1 ENST00000530511.1 |
POLD3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chr15_+_80364901 | 0.22 |
ENST00000560228.1
ENST00000559835.1 ENST00000559775.1 ENST00000558688.1 ENST00000560392.1 ENST00000560976.1 ENST00000558272.1 ENST00000558390.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr21_+_35552978 | 0.22 |
ENST00000428914.2
ENST00000609062.1 ENST00000609947.1 |
LINC00310
|
long intergenic non-protein coding RNA 310 |
| chr6_-_138866823 | 0.21 |
ENST00000342260.5
|
NHSL1
|
NHS-like 1 |
| chr12_-_10022735 | 0.21 |
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chrX_+_100645812 | 0.21 |
ENST00000427805.2
ENST00000553110.3 ENST00000392994.3 ENST00000409338.1 ENST00000409170.3 |
RPL36A
RPL36A-HNRNPH2
|
ribosomal protein L36a RPL36A-HNRNPH2 readthrough |
| chr2_+_109237717 | 0.21 |
ENST00000409441.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr20_-_5426380 | 0.21 |
ENST00000609252.1
ENST00000422352.1 |
LINC00658
|
long intergenic non-protein coding RNA 658 |
| chr4_+_54243798 | 0.21 |
ENST00000337488.6
ENST00000358575.5 ENST00000507922.1 |
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr16_-_11477562 | 0.21 |
ENST00000595168.1
|
CTD-3088G3.8
|
Protein LOC388210 |
| chr19_-_4535233 | 0.21 |
ENST00000381848.3
ENST00000588887.1 ENST00000586133.1 |
PLIN5
|
perilipin 5 |
| chr1_+_145525015 | 0.20 |
ENST00000539363.1
ENST00000538811.1 |
ITGA10
|
integrin, alpha 10 |
| chr2_+_132479948 | 0.20 |
ENST00000355171.4
|
C2orf27A
|
chromosome 2 open reading frame 27A |
| chr14_-_51027838 | 0.20 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr1_+_40942887 | 0.20 |
ENST00000372706.1
|
ZFP69
|
ZFP69 zinc finger protein |
| chr20_+_11873141 | 0.20 |
ENST00000422390.1
|
BTBD3
|
BTB (POZ) domain containing 3 |
| chr7_+_134576317 | 0.20 |
ENST00000424922.1
ENST00000495522.1 |
CALD1
|
caldesmon 1 |
| chr14_+_50291993 | 0.20 |
ENST00000595378.1
|
AL627171.2
|
HCG1786899; PRO2610; Uncharacterized protein |
| chr18_-_52626622 | 0.20 |
ENST00000591504.1
|
CCDC68
|
coiled-coil domain containing 68 |
| chr2_-_201729393 | 0.20 |
ENST00000321356.4
|
CLK1
|
CDC-like kinase 1 |
| chr12_-_11002063 | 0.19 |
ENST00000544994.1
ENST00000228811.4 ENST00000540107.1 |
PRR4
|
proline rich 4 (lacrimal) |
| chr14_+_56584414 | 0.19 |
ENST00000559044.1
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr4_+_156775910 | 0.19 |
ENST00000506072.1
ENST00000507590.1 |
TDO2
|
tryptophan 2,3-dioxygenase |
| chr15_+_63414760 | 0.19 |
ENST00000557972.1
|
LACTB
|
lactamase, beta |
| chr4_-_74853897 | 0.19 |
ENST00000296028.3
|
PPBP
|
pro-platelet basic protein (chemokine (C-X-C motif) ligand 7) |
| chr10_+_70480963 | 0.19 |
ENST00000265872.6
ENST00000535016.1 ENST00000538031.1 ENST00000543719.1 ENST00000539539.1 ENST00000543225.1 ENST00000536012.1 ENST00000494903.2 |
CCAR1
|
cell division cycle and apoptosis regulator 1 |
| chr11_-_82745238 | 0.19 |
ENST00000531021.1
|
RAB30
|
RAB30, member RAS oncogene family |
| chr9_+_135937365 | 0.18 |
ENST00000372080.4
ENST00000351304.7 |
CEL
|
carboxyl ester lipase |
| chr18_+_44526744 | 0.18 |
ENST00000585469.1
|
KATNAL2
|
katanin p60 subunit A-like 2 |
| chr14_+_20187174 | 0.18 |
ENST00000557414.1
|
OR4N2
|
olfactory receptor, family 4, subfamily N, member 2 |
| chr10_+_28822636 | 0.18 |
ENST00000442148.1
ENST00000448193.1 |
WAC
|
WW domain containing adaptor with coiled-coil |
| chr14_+_39703112 | 0.18 |
ENST00000555143.1
ENST00000280082.3 |
MIA2
|
melanoma inhibitory activity 2 |
| chr2_-_188312971 | 0.18 |
ENST00000410068.1
ENST00000447403.1 ENST00000410102.1 |
CALCRL
|
calcitonin receptor-like |
| chr5_+_81601166 | 0.18 |
ENST00000439350.1
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like |
| chr21_-_43786634 | 0.18 |
ENST00000291527.2
|
TFF1
|
trefoil factor 1 |
| chr8_+_32579321 | 0.18 |
ENST00000522402.1
|
NRG1
|
neuregulin 1 |
| chr5_-_156390230 | 0.18 |
ENST00000407087.3
ENST00000274532.2 |
TIMD4
|
T-cell immunoglobulin and mucin domain containing 4 |
| chr8_-_67940755 | 0.18 |
ENST00000324682.5
|
PPP1R42
|
protein phosphatase 1, regulatory subunit 42 |
| chr6_-_19804973 | 0.18 |
ENST00000457670.1
ENST00000607810.1 ENST00000606628.1 |
RP4-625H18.2
|
RP4-625H18.2 |
| chr11_-_85430163 | 0.18 |
ENST00000529581.1
ENST00000533577.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr3_+_171561127 | 0.18 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr4_+_102734967 | 0.18 |
ENST00000444316.2
|
BANK1
|
B-cell scaffold protein with ankyrin repeats 1 |
| chr4_+_74301880 | 0.18 |
ENST00000395792.2
ENST00000226359.2 |
AFP
|
alpha-fetoprotein |
| chr19_+_55105085 | 0.18 |
ENST00000251372.3
ENST00000453777.1 |
LILRA1
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 1 |
| chr3_-_101232019 | 0.17 |
ENST00000394095.2
ENST00000394091.1 ENST00000394094.2 ENST00000358203.3 ENST00000348610.3 ENST00000314261.7 |
SENP7
|
SUMO1/sentrin specific peptidase 7 |
| chr19_+_21265028 | 0.17 |
ENST00000291770.7
|
ZNF714
|
zinc finger protein 714 |
| chr6_+_76599809 | 0.17 |
ENST00000430435.1
|
MYO6
|
myosin VI |
| chr14_+_39944025 | 0.17 |
ENST00000554328.1
ENST00000556620.1 ENST00000557197.1 |
RP11-111A21.1
|
RP11-111A21.1 |
| chr15_+_63188009 | 0.17 |
ENST00000557900.1
|
RP11-1069G10.2
|
RP11-1069G10.2 |
| chr7_+_134576151 | 0.17 |
ENST00000393118.2
|
CALD1
|
caldesmon 1 |
| chr3_-_100566492 | 0.17 |
ENST00000528490.1
|
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr1_+_229440129 | 0.17 |
ENST00000366688.3
|
SPHAR
|
S-phase response (cyclin related) |
| chr6_+_31165901 | 0.17 |
ENST00000424675.1
|
HCG27
|
HLA complex group 27 (non-protein coding) |
| chr8_-_13134045 | 0.17 |
ENST00000512044.2
|
DLC1
|
deleted in liver cancer 1 |
| chr1_+_66797687 | 0.16 |
ENST00000371045.5
ENST00000531025.1 ENST00000526197.1 |
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chrM_+_3299 | 0.16 |
ENST00000361390.2
|
MT-ND1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr4_+_75174204 | 0.16 |
ENST00000332112.4
ENST00000514968.1 ENST00000503098.1 ENST00000502358.1 ENST00000509145.1 ENST00000505212.1 |
EPGN
|
epithelial mitogen |
| chr1_-_247171347 | 0.16 |
ENST00000339986.7
ENST00000487338.2 |
ZNF695
|
zinc finger protein 695 |
| chr5_-_75013193 | 0.16 |
ENST00000514838.2
ENST00000506164.1 ENST00000502826.1 ENST00000503835.1 ENST00000428202.2 ENST00000380475.2 |
POC5
|
POC5 centriolar protein |
| chr18_+_44526786 | 0.16 |
ENST00000245121.5
ENST00000356157.7 |
KATNAL2
|
katanin p60 subunit A-like 2 |
| chr7_+_64254793 | 0.16 |
ENST00000494380.1
ENST00000440155.2 ENST00000440598.1 ENST00000437743.1 |
ZNF138
|
zinc finger protein 138 |
| chr6_-_121655593 | 0.16 |
ENST00000398212.2
|
TBC1D32
|
TBC1 domain family, member 32 |
| chr12_-_91505608 | 0.16 |
ENST00000266718.4
|
LUM
|
lumican |
| chr21_+_17566643 | 0.16 |
ENST00000419952.1
ENST00000445461.2 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr14_-_38036271 | 0.16 |
ENST00000556024.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr10_+_91461337 | 0.16 |
ENST00000260753.4
ENST00000416354.1 ENST00000394289.2 ENST00000371728.3 |
KIF20B
|
kinesin family member 20B |
| chr3_+_111717511 | 0.16 |
ENST00000478951.1
ENST00000393917.2 |
TAGLN3
|
transgelin 3 |
| chr18_-_57027194 | 0.15 |
ENST00000251047.5
|
LMAN1
|
lectin, mannose-binding, 1 |
| chr6_-_10115007 | 0.15 |
ENST00000485268.1
|
OFCC1
|
orofacial cleft 1 candidate 1 |
| chr2_-_25100893 | 0.15 |
ENST00000433852.1
|
ADCY3
|
adenylate cyclase 3 |
| chr10_+_105726862 | 0.15 |
ENST00000335753.4
ENST00000369755.3 |
SLK
|
STE20-like kinase |
| chr2_-_61245363 | 0.15 |
ENST00000316752.6
|
PUS10
|
pseudouridylate synthase 10 |
| chr2_+_109204909 | 0.15 |
ENST00000393310.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr11_-_74204692 | 0.15 |
ENST00000528085.1
|
LIPT2
|
lipoyl(octanoyl) transferase 2 (putative) |
| chr11_-_96123022 | 0.15 |
ENST00000542662.1
|
CCDC82
|
coiled-coil domain containing 82 |
| chr3_+_98699880 | 0.15 |
ENST00000473756.1
|
LINC00973
|
long intergenic non-protein coding RNA 973 |
| chr2_-_201599892 | 0.15 |
ENST00000452787.1
|
AC007163.3
|
AC007163.3 |
| chr5_-_76788134 | 0.15 |
ENST00000507029.1
|
WDR41
|
WD repeat domain 41 |
| chr1_-_54411240 | 0.15 |
ENST00000371378.2
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr11_+_34654011 | 0.15 |
ENST00000531794.1
|
EHF
|
ets homologous factor |
| chr5_-_78808617 | 0.15 |
ENST00000282260.6
ENST00000508576.1 ENST00000535690.1 |
HOMER1
|
homer homolog 1 (Drosophila) |
| chr4_+_130017268 | 0.15 |
ENST00000425929.1
ENST00000508673.1 ENST00000508622.1 |
C4orf33
|
chromosome 4 open reading frame 33 |
| chr1_+_42928945 | 0.15 |
ENST00000428554.2
|
CCDC30
|
coiled-coil domain containing 30 |
| chr10_+_91461413 | 0.15 |
ENST00000447580.1
|
KIF20B
|
kinesin family member 20B |
| chr4_-_141348763 | 0.15 |
ENST00000509477.1
|
CLGN
|
calmegin |
| chr1_+_11072696 | 0.15 |
ENST00000240185.3
ENST00000476201.1 |
TARDBP
|
TAR DNA binding protein |
| chr22_+_35653445 | 0.14 |
ENST00000420166.1
ENST00000444518.2 ENST00000455359.1 ENST00000216106.5 |
HMGXB4
|
HMG box domain containing 4 |
| chr5_+_179135246 | 0.14 |
ENST00000508787.1
|
CANX
|
calnexin |
| chr1_+_207943667 | 0.14 |
ENST00000462968.2
|
CD46
|
CD46 molecule, complement regulatory protein |
| chr17_-_10633535 | 0.14 |
ENST00000341871.3
|
TMEM220
|
transmembrane protein 220 |
| chrY_+_15016013 | 0.14 |
ENST00000360160.4
ENST00000454054.1 |
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr4_+_11470867 | 0.14 |
ENST00000515343.1
|
RP11-281P23.1
|
RP11-281P23.1 |
| chr10_+_27793197 | 0.14 |
ENST00000356940.6
ENST00000535776.1 |
RAB18
|
RAB18, member RAS oncogene family |
| chr18_-_44500101 | 0.14 |
ENST00000589917.1
ENST00000587810.1 |
PIAS2
|
protein inhibitor of activated STAT, 2 |
| chr15_-_80263506 | 0.14 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr10_+_28822417 | 0.14 |
ENST00000428935.1
ENST00000420266.1 |
WAC
|
WW domain containing adaptor with coiled-coil |
| chr1_-_54411255 | 0.14 |
ENST00000371377.3
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr1_-_153433120 | 0.14 |
ENST00000368723.3
|
S100A7
|
S100 calcium binding protein A7 |
| chr16_+_16434185 | 0.14 |
ENST00000524823.2
|
AC138969.4
|
Protein PKD1P1 |
| chr2_-_201753717 | 0.14 |
ENST00000409264.2
|
PPIL3
|
peptidylprolyl isomerase (cyclophilin)-like 3 |
| chr16_+_33629600 | 0.14 |
ENST00000562905.2
|
IGHV3OR16-13
|
immunoglobulin heavy variable 3/OR16-13 (non-functional) |
| chr9_+_70971815 | 0.14 |
ENST00000396392.1
ENST00000396396.1 |
PGM5
|
phosphoglucomutase 5 |
| chr15_-_38519066 | 0.14 |
ENST00000561320.1
ENST00000561161.1 |
RP11-346D14.1
|
RP11-346D14.1 |
| chr3_-_57233966 | 0.14 |
ENST00000473921.1
ENST00000295934.3 |
HESX1
|
HESX homeobox 1 |
| chr7_+_64254766 | 0.14 |
ENST00000307355.7
ENST00000359735.3 |
ZNF138
|
zinc finger protein 138 |
| chr2_-_151344172 | 0.14 |
ENST00000375734.2
ENST00000263895.4 ENST00000454202.1 |
RND3
|
Rho family GTPase 3 |
| chrX_-_19688475 | 0.14 |
ENST00000541422.1
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr14_-_71107921 | 0.14 |
ENST00000553982.1
ENST00000500016.1 |
CTD-2540L5.5
CTD-2540L5.6
|
CTD-2540L5.5 CTD-2540L5.6 |
| chr2_-_45795145 | 0.14 |
ENST00000535761.1
|
SRBD1
|
S1 RNA binding domain 1 |
| chr7_-_99277610 | 0.13 |
ENST00000343703.5
ENST00000222982.4 ENST00000439761.1 ENST00000339843.2 |
CYP3A5
|
cytochrome P450, family 3, subfamily A, polypeptide 5 |
| chr8_+_32579341 | 0.13 |
ENST00000519240.1
ENST00000539990.1 |
NRG1
|
neuregulin 1 |
| chr6_-_112408661 | 0.13 |
ENST00000368662.5
|
TUBE1
|
tubulin, epsilon 1 |
| chr8_-_56987061 | 0.13 |
ENST00000009589.3
ENST00000524349.1 ENST00000519606.1 ENST00000519807.1 ENST00000521262.1 ENST00000520627.1 |
RPS20
|
ribosomal protein S20 |
| chr6_+_42749759 | 0.13 |
ENST00000314073.5
|
GLTSCR1L
|
GLTSCR1-like |
| chr20_-_10414804 | 0.13 |
ENST00000347364.3
|
MKKS
|
McKusick-Kaufman syndrome |
| chr8_-_123139423 | 0.13 |
ENST00000523792.1
|
RP11-398G24.2
|
RP11-398G24.2 |
| chr13_+_111893533 | 0.13 |
ENST00000478679.1
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr16_+_28663555 | 0.13 |
ENST00000526428.1
|
NPIPB8
|
nuclear pore complex interacting protein family, member B8 |
| chr10_+_86184676 | 0.13 |
ENST00000543283.1
|
CCSER2
|
coiled-coil serine-rich protein 2 |
| chr19_+_41281282 | 0.13 |
ENST00000263369.3
|
MIA
|
melanoma inhibitory activity |
| chr16_+_82068585 | 0.13 |
ENST00000563491.1
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr6_-_150212029 | 0.13 |
ENST00000529948.1
ENST00000357183.4 ENST00000367363.3 |
RAET1E
|
retinoic acid early transcript 1E |
| chr5_+_138678131 | 0.13 |
ENST00000394795.2
ENST00000510080.1 |
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr11_-_102323740 | 0.13 |
ENST00000398136.2
|
TMEM123
|
transmembrane protein 123 |
| chr2_-_162931052 | 0.13 |
ENST00000360534.3
|
DPP4
|
dipeptidyl-peptidase 4 |
| chr13_-_48575401 | 0.13 |
ENST00000433022.1
ENST00000544100.1 |
SUCLA2
|
succinate-CoA ligase, ADP-forming, beta subunit |
| chr7_+_116660246 | 0.13 |
ENST00000434836.1
ENST00000393443.1 ENST00000465133.1 ENST00000477742.1 ENST00000393447.4 ENST00000393444.3 |
ST7
|
suppression of tumorigenicity 7 |
| chr1_-_150669604 | 0.13 |
ENST00000427665.1
ENST00000540514.1 |
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr8_-_112248400 | 0.13 |
ENST00000519506.1
ENST00000522778.1 |
RP11-946L20.4
|
RP11-946L20.4 |
| chrX_-_154250989 | 0.13 |
ENST00000360256.4
|
F8
|
coagulation factor VIII, procoagulant component |
| chr1_+_145883868 | 0.13 |
ENST00000447947.2
|
GPR89C
|
G protein-coupled receptor 89C |
| chr12_-_111926342 | 0.13 |
ENST00000389154.3
|
ATXN2
|
ataxin 2 |
| chr1_-_54483842 | 0.13 |
ENST00000371362.3
ENST00000420619.1 |
LDLRAD1
|
low density lipoprotein receptor class A domain containing 1 |
| chr3_+_14693247 | 0.13 |
ENST00000383794.3
ENST00000303688.7 |
CCDC174
|
coiled-coil domain containing 174 |
| chr7_-_122342966 | 0.12 |
ENST00000447240.1
|
RNF148
|
ring finger protein 148 |
| chr14_+_35747825 | 0.12 |
ENST00000540871.1
|
PSMA6
|
proteasome (prosome, macropain) subunit, alpha type, 6 |
| chr4_+_70916119 | 0.12 |
ENST00000246896.3
ENST00000511674.1 |
HTN1
|
histatin 1 |
| chr5_+_110073853 | 0.12 |
ENST00000513807.1
ENST00000509442.2 |
SLC25A46
|
solute carrier family 25, member 46 |
| chr19_-_4717835 | 0.12 |
ENST00000599248.1
|
DPP9
|
dipeptidyl-peptidase 9 |
| chr2_-_110962544 | 0.12 |
ENST00000355301.4
ENST00000445609.2 ENST00000417665.1 ENST00000418527.1 ENST00000316534.4 ENST00000393272.3 |
NPHP1
|
nephronophthisis 1 (juvenile) |
| chr5_+_94890778 | 0.12 |
ENST00000380009.4
|
ARSK
|
arylsulfatase family, member K |
| chr11_+_74303575 | 0.12 |
ENST00000263681.2
|
POLD3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chrX_+_108779870 | 0.12 |
ENST00000372107.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr20_-_5426332 | 0.12 |
ENST00000420529.1
|
LINC00658
|
long intergenic non-protein coding RNA 658 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX13_SOX12
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0032714 | negative regulation of interleukin-5 production(GO:0032714) |
| 0.1 | 0.4 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.1 | 0.8 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.1 | 0.3 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.1 | 0.2 | GO:0071301 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.1 | 0.2 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.1 | 0.9 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 0.2 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.1 | 0.3 | GO:1903490 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.1 | 0.2 | GO:0035548 | gamma-delta T cell activation involved in immune response(GO:0002290) negative regulation of interferon-beta secretion(GO:0035548) regulation of gamma-delta T cell activation involved in immune response(GO:2001191) positive regulation of gamma-delta T cell activation involved in immune response(GO:2001193) |
| 0.0 | 0.2 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.8 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.2 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.1 | GO:0070101 | positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.0 | 0.3 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.2 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.1 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 0.2 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.0 | 0.2 | GO:0060179 | male mating behavior(GO:0060179) |
| 0.0 | 0.1 | GO:0006173 | dADP biosynthetic process(GO:0006173) |
| 0.0 | 0.1 | GO:1990426 | homologous recombination-dependent replication fork processing(GO:1990426) |
| 0.0 | 0.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.3 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.1 | GO:0071030 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.0 | 0.1 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.0 | 0.1 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.1 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.9 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.1 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
| 0.0 | 0.1 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.0 | 0.1 | GO:1904344 | positive regulation of growth rate(GO:0040010) regulation of gastric mucosal blood circulation(GO:1904344) positive regulation of gastric mucosal blood circulation(GO:1904346) gastric mucosal blood circulation(GO:1990768) |
| 0.0 | 0.2 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.0 | 0.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.2 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.2 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.0 | 0.7 | GO:0071624 | positive regulation of granulocyte chemotaxis(GO:0071624) |
| 0.0 | 0.1 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.0 | 0.1 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.1 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.1 | GO:0036343 | negative regulation of extracellular matrix disassembly(GO:0010716) psychomotor behavior(GO:0036343) |
| 0.0 | 0.1 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.0 | 0.1 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.0 | 0.1 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.0 | 0.1 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) negative regulation of transcription by transcription factor localization(GO:0010621) ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.0 | 0.1 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.0 | 0.1 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.0 | 0.4 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:0001835 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.1 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.1 | GO:0042245 | RNA repair(GO:0042245) |
| 0.0 | 0.5 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.1 | GO:0046035 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.2 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 0.2 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.1 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.1 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.1 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.0 | 0.6 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.1 | GO:0055095 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.0 | 0.1 | GO:0035750 | protein localization to myelin sheath abaxonal region(GO:0035750) |
| 0.0 | 0.2 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.1 | GO:0048749 | compound eye development(GO:0048749) |
| 0.0 | 0.1 | GO:2000230 | regulation of cholesterol transporter activity(GO:0060694) negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.0 | 0.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:1904550 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.0 | 0.0 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.1 | GO:1904977 | lymphatic endothelial cell migration(GO:1904977) |
| 0.0 | 0.2 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.0 | GO:0035377 | transepithelial water transport(GO:0035377) |
| 0.0 | 0.4 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.0 | 0.1 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.2 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.2 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.2 | GO:1900165 | negative regulation of interleukin-6 secretion(GO:1900165) |
| 0.0 | 0.2 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.2 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.2 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.1 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.1 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.3 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.1 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.1 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.1 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.1 | GO:0003051 | brain renin-angiotensin system(GO:0002035) angiotensin-mediated drinking behavior(GO:0003051) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.1 | GO:0044130 | negative regulation of growth of symbiont in host(GO:0044130) negative regulation of growth of symbiont involved in interaction with host(GO:0044146) |
| 0.0 | 0.0 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.0 | 0.0 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.2 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.0 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 0.0 | 0.1 | GO:2001270 | regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.2 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.0 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.1 | 0.3 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 0.3 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.0 | 0.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.1 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.8 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.5 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.3 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.3 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.0 | 0.4 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.2 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.1 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.0 | 0.2 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.8 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.4 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:0030430 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 0.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.3 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.9 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.1 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.1 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.1 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.3 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.2 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.0 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.1 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.0 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.0 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.1 | 0.8 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.2 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 0.1 | 0.2 | GO:0030107 | HLA-A specific inhibitory MHC class I receptor activity(GO:0030107) HLA-B specific inhibitory MHC class I receptor activity(GO:0030109) inhibitory MHC class I receptor activity(GO:0032396) |
| 0.0 | 0.5 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.4 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.4 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.3 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.2 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.0 | 0.3 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.2 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.1 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.0 | 0.1 | GO:0031768 | growth hormone-releasing hormone activity(GO:0016608) ghrelin receptor binding(GO:0031768) |
| 0.0 | 0.1 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 0.4 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.1 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 0.1 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.2 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.4 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.3 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.1 | GO:0044594 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.0 | 0.2 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 0.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.1 | GO:0035240 | dopamine binding(GO:0035240) |
| 0.0 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.1 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0008670 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.0 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.0 | 0.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.2 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.0 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.0 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.0 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 0.0 | 0.0 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.0 | 0.2 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.1 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.1 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.2 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |