Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
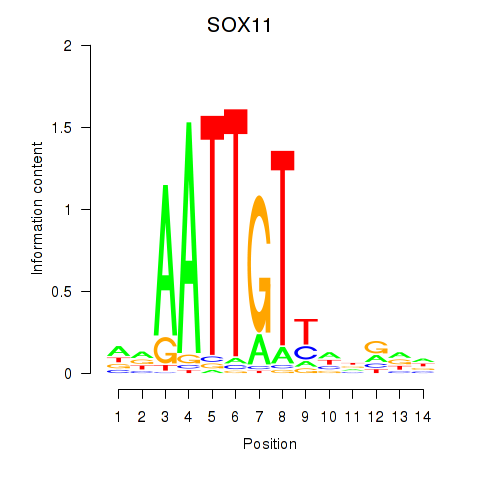
Results for SOX11
Z-value: 0.43
Transcription factors associated with SOX11
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX11
|
ENSG00000176887.5 | SRY-box transcription factor 11 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX11 | hg19_v2_chr2_+_5832799_5832799 | 0.11 | 8.9e-01 | Click! |
Activity profile of SOX11 motif
Sorted Z-values of SOX11 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_39989535 | 0.22 |
ENST00000356433.5
|
DLL3
|
delta-like 3 (Drosophila) |
| chr6_+_26183958 | 0.22 |
ENST00000356530.3
|
HIST1H2BE
|
histone cluster 1, H2be |
| chr6_-_154751629 | 0.20 |
ENST00000424998.1
|
CNKSR3
|
CNKSR family member 3 |
| chr2_-_176046391 | 0.18 |
ENST00000392541.3
ENST00000409194.1 |
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr2_-_37068530 | 0.16 |
ENST00000593798.1
|
AC007382.1
|
Uncharacterized protein |
| chr7_-_56118981 | 0.15 |
ENST00000419984.2
ENST00000413218.1 ENST00000424596.1 |
PSPH
|
phosphoserine phosphatase |
| chr10_+_5238793 | 0.15 |
ENST00000263126.1
|
AKR1C4
|
aldo-keto reductase family 1, member C4 |
| chr3_-_167191814 | 0.15 |
ENST00000466903.1
ENST00000264677.4 |
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2 |
| chr19_+_50094866 | 0.14 |
ENST00000418929.2
|
PRR12
|
proline rich 12 |
| chr16_+_67261008 | 0.14 |
ENST00000304800.9
ENST00000563953.1 ENST00000565201.1 |
TMEM208
|
transmembrane protein 208 |
| chrM_+_8366 | 0.14 |
ENST00000361851.1
|
MT-ATP8
|
mitochondrially encoded ATP synthase 8 |
| chr11_+_92085262 | 0.13 |
ENST00000298047.6
ENST00000409404.2 ENST00000541502.1 |
FAT3
|
FAT atypical cadherin 3 |
| chr20_-_30539773 | 0.13 |
ENST00000202017.4
|
PDRG1
|
p53 and DNA-damage regulated 1 |
| chr3_-_47950745 | 0.13 |
ENST00000429422.1
|
MAP4
|
microtubule-associated protein 4 |
| chr13_-_45048386 | 0.12 |
ENST00000472477.1
|
TSC22D1
|
TSC22 domain family, member 1 |
| chr17_+_6347761 | 0.12 |
ENST00000250056.8
ENST00000571373.1 ENST00000570337.2 ENST00000572595.2 ENST00000576056.1 |
FAM64A
|
family with sequence similarity 64, member A |
| chr4_-_105416039 | 0.12 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr11_-_64684672 | 0.12 |
ENST00000377264.3
ENST00000421419.2 |
ATG2A
|
autophagy related 2A |
| chr3_-_49967292 | 0.12 |
ENST00000455683.2
|
MON1A
|
MON1 secretory trafficking family member A |
| chr17_-_79895154 | 0.11 |
ENST00000405481.4
ENST00000585215.1 ENST00000577624.1 ENST00000403172.4 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chr15_-_36544450 | 0.11 |
ENST00000561394.1
|
RP11-184D12.1
|
RP11-184D12.1 |
| chr12_+_70574088 | 0.11 |
ENST00000552324.1
|
RP11-320P7.2
|
RP11-320P7.2 |
| chr7_-_143579973 | 0.11 |
ENST00000460532.1
ENST00000491908.1 |
FAM115A
|
family with sequence similarity 115, member A |
| chr17_-_79895097 | 0.10 |
ENST00000402252.2
ENST00000583564.1 ENST00000585244.1 ENST00000337943.5 ENST00000579698.1 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chr15_-_64673630 | 0.10 |
ENST00000558008.1
ENST00000559519.1 ENST00000380258.2 |
KIAA0101
|
KIAA0101 |
| chr15_-_64673665 | 0.10 |
ENST00000300035.4
|
KIAA0101
|
KIAA0101 |
| chr6_+_30585486 | 0.10 |
ENST00000259873.4
ENST00000506373.2 |
MRPS18B
|
mitochondrial ribosomal protein S18B |
| chr1_+_149754227 | 0.10 |
ENST00000444948.1
ENST00000369168.4 |
FCGR1A
|
Fc fragment of IgG, high affinity Ia, receptor (CD64) |
| chr17_-_72772462 | 0.10 |
ENST00000582870.1
ENST00000581136.1 ENST00000357814.3 ENST00000579218.1 ENST00000583476.1 ENST00000580301.1 ENST00000583757.1 ENST00000582524.1 |
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr4_+_71019903 | 0.10 |
ENST00000344526.5
|
C4orf40
|
chromosome 4 open reading frame 40 |
| chr11_-_62476694 | 0.10 |
ENST00000524862.1
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr2_-_37501692 | 0.09 |
ENST00000443977.1
|
PRKD3
|
protein kinase D3 |
| chr12_-_11287243 | 0.09 |
ENST00000539585.1
|
TAS2R30
|
taste receptor, type 2, member 30 |
| chr8_-_37707356 | 0.09 |
ENST00000520601.1
ENST00000521170.1 ENST00000220659.6 |
BRF2
|
BRF2, RNA polymerase III transcription initiation factor 50 kDa subunit |
| chr10_-_5446786 | 0.09 |
ENST00000479328.1
ENST00000380419.3 |
TUBAL3
|
tubulin, alpha-like 3 |
| chr20_-_32308028 | 0.09 |
ENST00000409299.3
ENST00000217398.3 ENST00000344022.3 |
PXMP4
|
peroxisomal membrane protein 4, 24kDa |
| chrX_-_7895755 | 0.08 |
ENST00000444736.1
ENST00000537427.1 ENST00000442940.1 |
PNPLA4
|
patatin-like phospholipase domain containing 4 |
| chr6_-_31125850 | 0.08 |
ENST00000507751.1
ENST00000448162.2 ENST00000502557.1 ENST00000503420.1 ENST00000507892.1 ENST00000507226.1 ENST00000513222.1 ENST00000503934.1 ENST00000396263.2 ENST00000508683.1 ENST00000428174.1 ENST00000448141.2 ENST00000507829.1 ENST00000455279.2 ENST00000376266.5 |
CCHCR1
|
coiled-coil alpha-helical rod protein 1 |
| chr4_+_57276661 | 0.08 |
ENST00000598320.1
|
AC068620.1
|
Uncharacterized protein |
| chr3_+_112930373 | 0.08 |
ENST00000498710.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr1_+_149239529 | 0.08 |
ENST00000457216.2
|
RP11-403I13.4
|
RP11-403I13.4 |
| chr21_+_43619796 | 0.08 |
ENST00000398457.2
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr2_+_105050794 | 0.08 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr17_+_6347729 | 0.08 |
ENST00000572447.1
|
FAM64A
|
family with sequence similarity 64, member A |
| chr3_+_112930387 | 0.07 |
ENST00000485230.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr5_+_140213815 | 0.07 |
ENST00000525929.1
ENST00000378125.3 |
PCDHA7
|
protocadherin alpha 7 |
| chr5_+_175085033 | 0.07 |
ENST00000377291.2
|
HRH2
|
histamine receptor H2 |
| chr2_-_190044480 | 0.07 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr17_+_38474489 | 0.07 |
ENST00000394089.2
ENST00000425707.3 |
RARA
|
retinoic acid receptor, alpha |
| chr6_+_131958436 | 0.07 |
ENST00000357639.3
ENST00000543135.1 ENST00000427148.2 ENST00000358229.5 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr6_-_170893268 | 0.06 |
ENST00000538195.1
|
PDCD2
|
programmed cell death 2 |
| chr1_-_219615984 | 0.06 |
ENST00000420762.1
|
RP11-95P13.1
|
RP11-95P13.1 |
| chr15_+_99433570 | 0.06 |
ENST00000558898.1
|
IGF1R
|
insulin-like growth factor 1 receptor |
| chrX_-_109590174 | 0.06 |
ENST00000372054.1
|
GNG5P2
|
guanine nucleotide binding protein (G protein), gamma 5 pseudogene 2 |
| chr2_+_42104692 | 0.06 |
ENST00000398796.2
ENST00000442214.1 |
AC104654.1
|
AC104654.1 |
| chr2_-_219157250 | 0.06 |
ENST00000434015.2
ENST00000444183.1 ENST00000420341.1 ENST00000453281.1 ENST00000258412.3 ENST00000440422.1 |
TMBIM1
|
transmembrane BAX inhibitor motif containing 1 |
| chr17_+_61151306 | 0.06 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr4_-_159080806 | 0.06 |
ENST00000590648.1
|
FAM198B
|
family with sequence similarity 198, member B |
| chr9_+_12775011 | 0.06 |
ENST00000319264.3
|
LURAP1L
|
leucine rich adaptor protein 1-like |
| chr7_+_90032821 | 0.05 |
ENST00000427904.1
|
CLDN12
|
claudin 12 |
| chr14_+_21156915 | 0.05 |
ENST00000397990.4
ENST00000555597.1 |
ANG
RNASE4
|
angiogenin, ribonuclease, RNase A family, 5 ribonuclease, RNase A family, 4 |
| chr12_-_122240792 | 0.05 |
ENST00000545885.1
ENST00000542933.1 ENST00000428029.2 ENST00000541694.1 ENST00000536662.1 ENST00000535643.1 ENST00000541657.1 |
AC084018.1
RHOF
|
AC084018.1 ras homolog family member F (in filopodia) |
| chr2_+_166326157 | 0.05 |
ENST00000421875.1
ENST00000314499.7 ENST00000409664.1 |
CSRNP3
|
cysteine-serine-rich nuclear protein 3 |
| chr4_+_144312659 | 0.05 |
ENST00000509992.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr21_+_17792672 | 0.05 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr1_-_205744574 | 0.05 |
ENST00000367139.3
ENST00000235932.4 ENST00000437324.2 ENST00000414729.1 |
RAB7L1
|
RAB7, member RAS oncogene family-like 1 |
| chr1_+_196621156 | 0.05 |
ENST00000359637.2
|
CFH
|
complement factor H |
| chr8_-_623547 | 0.05 |
ENST00000522893.1
|
ERICH1
|
glutamate-rich 1 |
| chr2_+_138722028 | 0.05 |
ENST00000280096.5
|
HNMT
|
histamine N-methyltransferase |
| chr8_-_128960591 | 0.05 |
ENST00000539634.1
|
TMEM75
|
transmembrane protein 75 |
| chr17_-_66951474 | 0.05 |
ENST00000269080.2
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr8_-_25281747 | 0.05 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr15_+_45028719 | 0.04 |
ENST00000560442.1
ENST00000558329.1 ENST00000561043.1 |
TRIM69
|
tripartite motif containing 69 |
| chr5_+_140248518 | 0.04 |
ENST00000398640.2
|
PCDHA11
|
protocadherin alpha 11 |
| chr4_-_90759440 | 0.04 |
ENST00000336904.3
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr11_-_58611957 | 0.04 |
ENST00000532258.1
|
GLYATL2
|
glycine-N-acyltransferase-like 2 |
| chr17_+_47448102 | 0.04 |
ENST00000576461.1
|
RP11-81K2.1
|
Uncharacterized protein |
| chr2_-_71222466 | 0.04 |
ENST00000606025.1
|
AC007040.11
|
Uncharacterized protein |
| chr2_+_207804278 | 0.04 |
ENST00000272852.3
|
CPO
|
carboxypeptidase O |
| chr7_-_92855762 | 0.04 |
ENST00000453812.2
ENST00000394468.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr4_+_72204755 | 0.04 |
ENST00000512686.1
ENST00000340595.3 |
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chrX_+_107288197 | 0.03 |
ENST00000415430.3
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr18_+_46065393 | 0.03 |
ENST00000256413.3
|
CTIF
|
CBP80/20-dependent translation initiation factor |
| chr1_+_146373546 | 0.03 |
ENST00000446760.2
|
NBPF12
|
neuroblastoma breakpoint family, member 12 |
| chr22_+_24407776 | 0.03 |
ENST00000405822.2
|
CABIN1
|
calcineurin binding protein 1 |
| chr19_-_5680499 | 0.03 |
ENST00000587589.1
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr14_+_39703112 | 0.03 |
ENST00000555143.1
ENST00000280082.3 |
MIA2
|
melanoma inhibitory activity 2 |
| chr1_+_196621002 | 0.03 |
ENST00000367429.4
ENST00000439155.2 |
CFH
|
complement factor H |
| chr3_-_112564797 | 0.03 |
ENST00000398214.1
ENST00000448932.1 |
CD200R1L
|
CD200 receptor 1-like |
| chr5_+_140207536 | 0.03 |
ENST00000529310.1
ENST00000527624.1 |
PCDHA6
|
protocadherin alpha 6 |
| chr17_-_66951382 | 0.03 |
ENST00000586539.1
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr2_+_138721850 | 0.03 |
ENST00000329366.4
ENST00000280097.3 |
HNMT
|
histamine N-methyltransferase |
| chr13_+_76413852 | 0.03 |
ENST00000533809.2
|
LMO7
|
LIM domain 7 |
| chr5_-_115872124 | 0.03 |
ENST00000515009.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chrX_+_102192200 | 0.03 |
ENST00000218249.5
|
RAB40AL
|
RAB40A, member RAS oncogene family-like |
| chrX_+_107288239 | 0.03 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr4_-_186570679 | 0.02 |
ENST00000451974.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr2_+_27799389 | 0.02 |
ENST00000408964.2
|
C2orf16
|
chromosome 2 open reading frame 16 |
| chr15_+_80733570 | 0.02 |
ENST00000533983.1
ENST00000527771.1 ENST00000525103.1 |
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr13_-_30424821 | 0.02 |
ENST00000380680.4
|
UBL3
|
ubiquitin-like 3 |
| chr12_-_22063787 | 0.02 |
ENST00000544039.1
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr5_+_140753444 | 0.02 |
ENST00000517434.1
|
PCDHGA6
|
protocadherin gamma subfamily A, 6 |
| chr3_+_113251143 | 0.02 |
ENST00000264852.4
ENST00000393830.3 |
SIDT1
|
SID1 transmembrane family, member 1 |
| chr18_+_46065483 | 0.02 |
ENST00000382998.4
|
CTIF
|
CBP80/20-dependent translation initiation factor |
| chr9_+_75766763 | 0.02 |
ENST00000456643.1
ENST00000415424.1 |
ANXA1
|
annexin A1 |
| chr3_-_87325728 | 0.02 |
ENST00000350375.2
|
POU1F1
|
POU class 1 homeobox 1 |
| chr10_+_32873190 | 0.02 |
ENST00000375025.4
|
C10orf68
|
Homo sapiens coiled-coil domain containing 7 (CCDC7), transcript variant 5, mRNA. |
| chr1_-_205744205 | 0.02 |
ENST00000446390.2
|
RAB7L1
|
RAB7, member RAS oncogene family-like 1 |
| chr7_+_98923505 | 0.02 |
ENST00000432884.2
ENST00000262942.5 |
ARPC1A
|
actin related protein 2/3 complex, subunit 1A, 41kDa |
| chr4_-_110624564 | 0.02 |
ENST00000352981.3
ENST00000265164.2 ENST00000505486.1 |
CASP6
|
caspase 6, apoptosis-related cysteine peptidase |
| chr8_+_104831472 | 0.02 |
ENST00000262231.10
ENST00000507740.1 |
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr19_-_6501778 | 0.02 |
ENST00000596291.1
|
TUBB4A
|
tubulin, beta 4A class IVa |
| chr3_-_18466026 | 0.02 |
ENST00000417717.2
|
SATB1
|
SATB homeobox 1 |
| chr3_+_101504200 | 0.02 |
ENST00000422132.1
|
NXPE3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr6_-_56492816 | 0.02 |
ENST00000522360.1
|
DST
|
dystonin |
| chr5_-_115872142 | 0.02 |
ENST00000510263.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chrX_-_100548045 | 0.02 |
ENST00000372907.3
ENST00000372905.2 |
TAF7L
|
TAF7-like RNA polymerase II, TATA box binding protein (TBP)-associated factor, 50kDa |
| chr15_-_78913628 | 0.02 |
ENST00000348639.3
|
CHRNA3
|
cholinergic receptor, nicotinic, alpha 3 (neuronal) |
| chr9_-_95166841 | 0.02 |
ENST00000262551.4
|
OGN
|
osteoglycin |
| chr17_+_35732916 | 0.02 |
ENST00000586700.1
|
C17orf78
|
chromosome 17 open reading frame 78 |
| chr3_+_112930306 | 0.02 |
ENST00000495514.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr12_+_56075330 | 0.02 |
ENST00000394252.3
|
METTL7B
|
methyltransferase like 7B |
| chr17_-_10276319 | 0.01 |
ENST00000252172.4
ENST00000418404.3 |
MYH13
|
myosin, heavy chain 13, skeletal muscle |
| chr6_-_11779014 | 0.01 |
ENST00000229583.5
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr7_-_7575477 | 0.01 |
ENST00000399429.3
|
COL28A1
|
collagen, type XXVIII, alpha 1 |
| chr7_+_90012986 | 0.01 |
ENST00000416322.1
|
CLDN12
|
claudin 12 |
| chr11_+_22688150 | 0.01 |
ENST00000454584.2
|
GAS2
|
growth arrest-specific 2 |
| chr1_+_198608146 | 0.01 |
ENST00000367376.2
ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr10_+_69865866 | 0.01 |
ENST00000354393.2
|
MYPN
|
myopalladin |
| chr5_+_40841410 | 0.01 |
ENST00000381677.3
|
CARD6
|
caspase recruitment domain family, member 6 |
| chr17_-_72772425 | 0.01 |
ENST00000578822.1
|
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr10_-_127505167 | 0.01 |
ENST00000368786.1
|
UROS
|
uroporphyrinogen III synthase |
| chr2_+_102928009 | 0.01 |
ENST00000404917.2
ENST00000447231.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chrX_+_24483338 | 0.01 |
ENST00000379162.4
ENST00000441463.2 |
PDK3
|
pyruvate dehydrogenase kinase, isozyme 3 |
| chr5_+_179159813 | 0.01 |
ENST00000292599.3
|
MAML1
|
mastermind-like 1 (Drosophila) |
| chr6_-_11779174 | 0.01 |
ENST00000379413.2
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr3_+_130279178 | 0.01 |
ENST00000358511.6
ENST00000453409.2 |
COL6A6
|
collagen, type VI, alpha 6 |
| chr11_-_82611448 | 0.01 |
ENST00000393399.2
ENST00000313010.3 |
PRCP
|
prolylcarboxypeptidase (angiotensinase C) |
| chrM_+_7586 | 0.01 |
ENST00000361739.1
|
MT-CO2
|
mitochondrially encoded cytochrome c oxidase II |
| chr22_-_30642728 | 0.01 |
ENST00000403987.3
|
LIF
|
leukemia inhibitory factor |
| chr2_-_20251744 | 0.01 |
ENST00000175091.4
|
LAPTM4A
|
lysosomal protein transmembrane 4 alpha |
| chr7_-_81399355 | 0.01 |
ENST00000457544.2
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr8_+_104831440 | 0.01 |
ENST00000515551.1
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr3_-_87325612 | 0.01 |
ENST00000561167.1
ENST00000560656.1 ENST00000344265.3 |
POU1F1
|
POU class 1 homeobox 1 |
| chr15_+_69857515 | 0.01 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr8_+_41386761 | 0.01 |
ENST00000523277.2
|
GINS4
|
GINS complex subunit 4 (Sld5 homolog) |
| chr15_-_37392086 | 0.01 |
ENST00000561208.1
|
MEIS2
|
Meis homeobox 2 |
| chr9_-_114521783 | 0.01 |
ENST00000394779.3
ENST00000394777.4 |
C9orf84
|
chromosome 9 open reading frame 84 |
| chr6_-_89927151 | 0.00 |
ENST00000454853.2
|
GABRR1
|
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
| chr2_-_73869508 | 0.00 |
ENST00000272425.3
|
NAT8
|
N-acetyltransferase 8 (GCN5-related, putative) |
| chrX_+_135388147 | 0.00 |
ENST00000394141.1
|
GPR112
|
G protein-coupled receptor 112 |
| chr13_+_78315295 | 0.00 |
ENST00000351546.3
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr4_-_22444733 | 0.00 |
ENST00000508133.1
|
GPR125
|
G protein-coupled receptor 125 |
| chr3_+_113616317 | 0.00 |
ENST00000440446.2
ENST00000488680.1 |
GRAMD1C
|
GRAM domain containing 1C |
| chr8_+_104831554 | 0.00 |
ENST00000408894.2
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr17_-_6524159 | 0.00 |
ENST00000589033.1
|
KIAA0753
|
KIAA0753 |
| chr12_-_772901 | 0.00 |
ENST00000305108.4
|
NINJ2
|
ninjurin 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX11
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.2 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.2 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.1 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.0 | GO:1990637 | response to prolactin(GO:1990637) regulation of ovarian follicle development(GO:2000354) |
| 0.0 | 0.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.1 | GO:2000504 | negative regulation of Fas signaling pathway(GO:1902045) positive regulation of blood vessel remodeling(GO:2000504) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.1 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.0 | 0.1 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.1 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.0 | 0.0 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0035529 | phosphodiesterase I activity(GO:0004528) NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:0019864 | IgG binding(GO:0019864) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |