Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
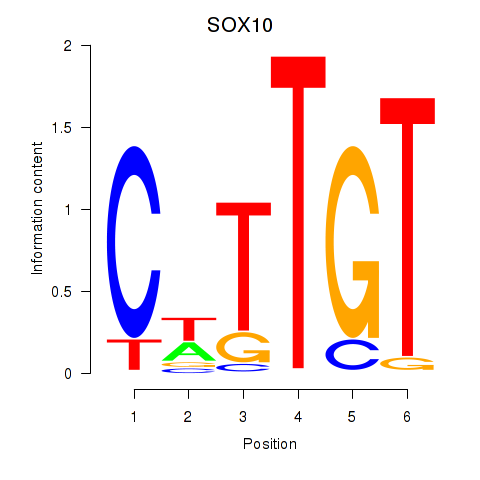
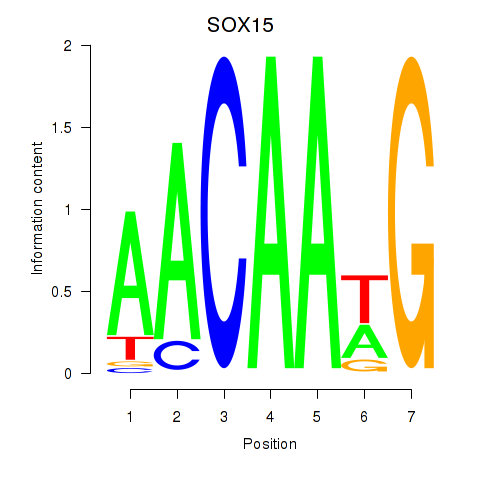
Results for SOX10_SOX15
Z-value: 0.70
Transcription factors associated with SOX10_SOX15
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX10
|
ENSG00000100146.12 | SRY-box transcription factor 10 |
|
SOX15
|
ENSG00000129194.3 | SRY-box transcription factor 15 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX15 | hg19_v2_chr17_-_7493390_7493488 | -0.78 | 2.2e-01 | Click! |
| SOX10 | hg19_v2_chr22_-_38380543_38380569 | -0.10 | 9.0e-01 | Click! |
Activity profile of SOX10_SOX15 motif
Sorted Z-values of SOX10_SOX15 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_76649753 | 0.83 |
ENST00000603759.1
|
USO1
|
USO1 vesicle transport factor |
| chr11_+_110001723 | 0.50 |
ENST00000528673.1
|
ZC3H12C
|
zinc finger CCCH-type containing 12C |
| chr12_+_95611536 | 0.43 |
ENST00000549002.1
|
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr14_-_23791484 | 0.42 |
ENST00000594872.1
|
AL049829.1
|
Uncharacterized protein |
| chr5_+_149865377 | 0.42 |
ENST00000522491.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr14_-_51027838 | 0.39 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr6_-_127840336 | 0.37 |
ENST00000525778.1
|
SOGA3
|
SOGA family member 3 |
| chr8_-_42358742 | 0.33 |
ENST00000517366.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr7_-_128415844 | 0.32 |
ENST00000249389.2
|
OPN1SW
|
opsin 1 (cone pigments), short-wave-sensitive |
| chr1_+_197886461 | 0.30 |
ENST00000367388.3
ENST00000337020.2 ENST00000367387.4 |
LHX9
|
LIM homeobox 9 |
| chr8_-_124553437 | 0.30 |
ENST00000517956.1
ENST00000443022.2 |
FBXO32
|
F-box protein 32 |
| chr6_-_127840021 | 0.29 |
ENST00000465909.2
|
SOGA3
|
SOGA family member 3 |
| chr7_-_27219849 | 0.29 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr2_-_118943930 | 0.28 |
ENST00000449075.1
ENST00000414886.1 ENST00000449819.1 |
AC093901.1
|
AC093901.1 |
| chr22_+_37959647 | 0.28 |
ENST00000415670.1
|
CDC42EP1
|
CDC42 effector protein (Rho GTPase binding) 1 |
| chr16_+_53242350 | 0.25 |
ENST00000565442.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr6_-_127840048 | 0.25 |
ENST00000467753.1
|
SOGA3
|
SOGA family member 3 |
| chr8_+_61822605 | 0.21 |
ENST00000526936.1
|
AC022182.1
|
AC022182.1 |
| chr17_-_39041479 | 0.20 |
ENST00000167588.3
|
KRT20
|
keratin 20 |
| chr4_-_140477910 | 0.19 |
ENST00000404104.3
|
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr7_+_23146271 | 0.19 |
ENST00000545771.1
|
KLHL7
|
kelch-like family member 7 |
| chr18_-_74207146 | 0.17 |
ENST00000443185.2
|
ZNF516
|
zinc finger protein 516 |
| chr3_+_111260980 | 0.17 |
ENST00000438817.2
|
CD96
|
CD96 molecule |
| chr5_+_150406527 | 0.17 |
ENST00000520059.1
|
GPX3
|
glutathione peroxidase 3 (plasma) |
| chr15_-_45459704 | 0.17 |
ENST00000558039.1
|
CTD-2651B20.1
|
CTD-2651B20.1 |
| chr10_-_75351088 | 0.16 |
ENST00000451492.1
ENST00000413442.1 |
USP54
|
ubiquitin specific peptidase 54 |
| chr1_+_201857798 | 0.15 |
ENST00000362011.6
|
SHISA4
|
shisa family member 4 |
| chr11_-_46141338 | 0.15 |
ENST00000529782.1
ENST00000532010.1 ENST00000525438.1 ENST00000533757.1 ENST00000527782.1 |
PHF21A
|
PHD finger protein 21A |
| chr14_-_91884150 | 0.15 |
ENST00000553403.1
|
CCDC88C
|
coiled-coil domain containing 88C |
| chr16_+_88636875 | 0.15 |
ENST00000569435.1
|
ZC3H18
|
zinc finger CCCH-type containing 18 |
| chr2_+_232573222 | 0.15 |
ENST00000341369.7
ENST00000409683.1 |
PTMA
|
prothymosin, alpha |
| chr5_-_138780159 | 0.15 |
ENST00000512473.1
ENST00000515581.1 ENST00000515277.1 |
DNAJC18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr2_-_165697717 | 0.14 |
ENST00000439313.1
|
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr2_-_153573965 | 0.14 |
ENST00000448428.1
|
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr14_-_50999190 | 0.14 |
ENST00000557390.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr11_-_130184470 | 0.14 |
ENST00000357899.4
ENST00000397753.1 |
ZBTB44
|
zinc finger and BTB domain containing 44 |
| chr5_-_65018834 | 0.14 |
ENST00000506816.1
|
SGTB
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr7_-_76247617 | 0.14 |
ENST00000441393.1
|
POMZP3
|
POM121 and ZP3 fusion |
| chr17_+_21730180 | 0.14 |
ENST00000584398.1
|
UBBP4
|
ubiquitin B pseudogene 4 |
| chr5_-_111092873 | 0.14 |
ENST00000509025.1
ENST00000515855.1 |
NREP
|
neuronal regeneration related protein |
| chr5_+_86563636 | 0.13 |
ENST00000274376.6
|
RASA1
|
RAS p21 protein activator (GTPase activating protein) 1 |
| chr2_+_120517717 | 0.13 |
ENST00000420482.1
ENST00000488279.2 |
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr17_-_77924627 | 0.13 |
ENST00000572862.1
ENST00000573782.1 ENST00000574427.1 ENST00000570373.1 ENST00000340848.7 ENST00000576768.1 |
TBC1D16
|
TBC1 domain family, member 16 |
| chr12_-_49504449 | 0.13 |
ENST00000547675.1
|
LMBR1L
|
limb development membrane protein 1-like |
| chr6_-_31632962 | 0.13 |
ENST00000456540.1
ENST00000445768.1 |
GPANK1
|
G patch domain and ankyrin repeats 1 |
| chr10_+_114710211 | 0.13 |
ENST00000349937.2
ENST00000369397.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr3_+_160473343 | 0.13 |
ENST00000497343.1
|
PPM1L
|
protein phosphatase, Mg2+/Mn2+ dependent, 1L |
| chr12_-_42631529 | 0.12 |
ENST00000548917.1
|
YAF2
|
YY1 associated factor 2 |
| chr2_+_149402989 | 0.12 |
ENST00000397424.2
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr17_-_79881408 | 0.12 |
ENST00000392366.3
|
MAFG
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog G |
| chr3_-_52719912 | 0.12 |
ENST00000420148.1
|
PBRM1
|
polybromo 1 |
| chr13_-_41593425 | 0.12 |
ENST00000239882.3
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chrY_+_15815447 | 0.12 |
ENST00000284856.3
|
TMSB4Y
|
thymosin beta 4, Y-linked |
| chr12_+_54378849 | 0.12 |
ENST00000515593.1
|
HOXC10
|
homeobox C10 |
| chr15_+_63335899 | 0.12 |
ENST00000561266.1
|
TPM1
|
tropomyosin 1 (alpha) |
| chrX_+_70503526 | 0.12 |
ENST00000413858.1
ENST00000450092.1 |
NONO
|
non-POU domain containing, octamer-binding |
| chr15_+_52043813 | 0.11 |
ENST00000435126.2
|
TMOD2
|
tropomodulin 2 (neuronal) |
| chr2_-_97534312 | 0.11 |
ENST00000442264.1
|
SEMA4C
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C |
| chr9_-_14308004 | 0.11 |
ENST00000493697.1
|
NFIB
|
nuclear factor I/B |
| chr12_-_2966193 | 0.11 |
ENST00000382678.3
|
AC005841.1
|
Uncharacterized protein ENSP00000372125 |
| chr14_-_80678512 | 0.11 |
ENST00000553968.1
|
DIO2
|
deiodinase, iodothyronine, type II |
| chr4_-_76649546 | 0.11 |
ENST00000508510.1
ENST00000509561.1 ENST00000499709.2 ENST00000511868.1 |
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr4_+_41614720 | 0.11 |
ENST00000509277.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr12_-_109797249 | 0.11 |
ENST00000538041.1
|
RP11-256L11.1
|
RP11-256L11.1 |
| chr6_+_155443048 | 0.11 |
ENST00000535583.1
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr18_+_3450161 | 0.10 |
ENST00000551402.1
ENST00000577543.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr1_+_93645314 | 0.10 |
ENST00000343253.7
|
CCDC18
|
coiled-coil domain containing 18 |
| chr22_+_19710468 | 0.10 |
ENST00000366425.3
|
GP1BB
|
glycoprotein Ib (platelet), beta polypeptide |
| chr6_+_30848740 | 0.10 |
ENST00000505534.1
|
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr2_+_232573208 | 0.10 |
ENST00000409115.3
|
PTMA
|
prothymosin, alpha |
| chr1_+_47799542 | 0.10 |
ENST00000471289.2
ENST00000450808.2 |
CMPK1
|
cytidine monophosphate (UMP-CMP) kinase 1, cytosolic |
| chr6_+_30848771 | 0.10 |
ENST00000503180.1
|
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr4_+_141677577 | 0.10 |
ENST00000609937.1
|
RP11-102N12.3
|
RP11-102N12.3 |
| chrX_+_78003204 | 0.10 |
ENST00000435339.3
ENST00000514744.1 |
LPAR4
|
lysophosphatidic acid receptor 4 |
| chr10_-_735553 | 0.09 |
ENST00000280886.6
ENST00000423550.1 |
DIP2C
|
DIP2 disco-interacting protein 2 homolog C (Drosophila) |
| chr1_+_93913713 | 0.09 |
ENST00000604705.1
ENST00000370253.2 |
FNBP1L
|
formin binding protein 1-like |
| chr5_-_38557561 | 0.09 |
ENST00000511561.1
|
LIFR
|
leukemia inhibitory factor receptor alpha |
| chr14_-_71107921 | 0.09 |
ENST00000553982.1
ENST00000500016.1 |
CTD-2540L5.5
CTD-2540L5.6
|
CTD-2540L5.5 CTD-2540L5.6 |
| chr22_+_31742875 | 0.09 |
ENST00000504184.2
|
AC005003.1
|
CDNA FLJ20464 fis, clone KAT06158; HCG1777549; Uncharacterized protein |
| chr16_+_53241854 | 0.09 |
ENST00000565803.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr11_-_94965667 | 0.08 |
ENST00000542176.1
ENST00000278499.2 |
SESN3
|
sestrin 3 |
| chrX_+_70503433 | 0.08 |
ENST00000276079.8
ENST00000373856.3 ENST00000373841.1 ENST00000420903.1 |
NONO
|
non-POU domain containing, octamer-binding |
| chr8_+_21777243 | 0.08 |
ENST00000521303.1
|
XPO7
|
exportin 7 |
| chr12_-_12419703 | 0.08 |
ENST00000543091.1
ENST00000261349.4 |
LRP6
|
low density lipoprotein receptor-related protein 6 |
| chr17_+_36861735 | 0.08 |
ENST00000378137.5
ENST00000325718.7 |
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6 |
| chr7_+_20370300 | 0.08 |
ENST00000537992.1
|
ITGB8
|
integrin, beta 8 |
| chr3_-_149688971 | 0.08 |
ENST00000498307.1
ENST00000489155.1 |
PFN2
|
profilin 2 |
| chr11_-_46848393 | 0.08 |
ENST00000526496.1
|
CKAP5
|
cytoskeleton associated protein 5 |
| chr7_-_105926058 | 0.08 |
ENST00000417537.1
|
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chr3_+_181429704 | 0.07 |
ENST00000431565.2
ENST00000325404.1 |
SOX2
|
SRY (sex determining region Y)-box 2 |
| chr13_-_108518986 | 0.07 |
ENST00000375915.2
|
FAM155A
|
family with sequence similarity 155, member A |
| chr7_+_69064566 | 0.07 |
ENST00000403018.2
|
AUTS2
|
autism susceptibility candidate 2 |
| chr3_-_123123407 | 0.07 |
ENST00000466617.1
|
ADCY5
|
adenylate cyclase 5 |
| chr6_+_108881012 | 0.07 |
ENST00000343882.6
|
FOXO3
|
forkhead box O3 |
| chrX_+_69674943 | 0.07 |
ENST00000542398.1
|
DLG3
|
discs, large homolog 3 (Drosophila) |
| chr19_-_19626838 | 0.07 |
ENST00000360913.3
|
TSSK6
|
testis-specific serine kinase 6 |
| chr5_+_140729649 | 0.07 |
ENST00000523390.1
|
PCDHGB1
|
protocadherin gamma subfamily B, 1 |
| chr17_+_67498538 | 0.07 |
ENST00000589647.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr7_+_20370746 | 0.07 |
ENST00000222573.4
|
ITGB8
|
integrin, beta 8 |
| chr4_+_41614909 | 0.07 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr9_-_123476612 | 0.07 |
ENST00000426959.1
|
MEGF9
|
multiple EGF-like-domains 9 |
| chr3_-_123339343 | 0.07 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr7_-_140178726 | 0.07 |
ENST00000480552.1
|
MKRN1
|
makorin ring finger protein 1 |
| chr6_-_161695042 | 0.07 |
ENST00000366908.5
ENST00000366911.5 ENST00000366905.3 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr12_+_27398584 | 0.07 |
ENST00000543246.1
|
STK38L
|
serine/threonine kinase 38 like |
| chr14_-_31495569 | 0.07 |
ENST00000357479.5
ENST00000355683.5 |
STRN3
|
striatin, calmodulin binding protein 3 |
| chr16_-_73082274 | 0.07 |
ENST00000268489.5
|
ZFHX3
|
zinc finger homeobox 3 |
| chrX_-_63425561 | 0.07 |
ENST00000374869.3
ENST00000330258.3 |
AMER1
|
APC membrane recruitment protein 1 |
| chr3_-_188665428 | 0.07 |
ENST00000444488.1
|
TPRG1-AS1
|
TPRG1 antisense RNA 1 |
| chr12_-_96793142 | 0.07 |
ENST00000552262.1
ENST00000551816.1 ENST00000552496.1 |
CDK17
|
cyclin-dependent kinase 17 |
| chr14_-_91884115 | 0.07 |
ENST00000389857.6
|
CCDC88C
|
coiled-coil domain containing 88C |
| chr5_-_58571935 | 0.07 |
ENST00000503258.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr14_+_38033252 | 0.07 |
ENST00000554829.1
|
RP11-356O9.1
|
RP11-356O9.1 |
| chr15_+_57211318 | 0.07 |
ENST00000557947.1
|
TCF12
|
transcription factor 12 |
| chr4_+_26323764 | 0.07 |
ENST00000514730.1
ENST00000507574.1 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr7_+_29234101 | 0.07 |
ENST00000435288.2
|
CHN2
|
chimerin 2 |
| chr7_-_15601595 | 0.07 |
ENST00000342526.3
|
AGMO
|
alkylglycerol monooxygenase |
| chr7_-_117512264 | 0.07 |
ENST00000454375.1
|
CTTNBP2
|
cortactin binding protein 2 |
| chr6_-_111888474 | 0.07 |
ENST00000368735.1
|
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chrX_-_128788914 | 0.06 |
ENST00000429967.1
ENST00000307484.6 |
APLN
|
apelin |
| chr2_-_70475586 | 0.06 |
ENST00000416149.2
|
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr18_-_45456693 | 0.06 |
ENST00000587421.1
|
SMAD2
|
SMAD family member 2 |
| chr1_+_183774240 | 0.06 |
ENST00000360851.3
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1 |
| chr17_+_29815013 | 0.06 |
ENST00000394744.2
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II) |
| chr5_-_137090028 | 0.06 |
ENST00000314940.4
|
HNRNPA0
|
heterogeneous nuclear ribonucleoprotein A0 |
| chr22_-_39150947 | 0.06 |
ENST00000411587.2
ENST00000420859.1 ENST00000452294.1 ENST00000456894.1 |
SUN2
|
Sad1 and UNC84 domain containing 2 |
| chr2_+_66662249 | 0.06 |
ENST00000560281.2
|
MEIS1
|
Meis homeobox 1 |
| chr3_-_62358690 | 0.06 |
ENST00000475839.1
|
FEZF2
|
FEZ family zinc finger 2 |
| chr5_-_114515734 | 0.06 |
ENST00000514154.1
ENST00000282369.3 |
TRIM36
|
tripartite motif containing 36 |
| chr9_-_123476719 | 0.06 |
ENST00000373930.3
|
MEGF9
|
multiple EGF-like-domains 9 |
| chr8_+_81397846 | 0.06 |
ENST00000379091.4
|
ZBTB10
|
zinc finger and BTB domain containing 10 |
| chr2_+_120517174 | 0.06 |
ENST00000263708.2
|
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr19_+_47105309 | 0.06 |
ENST00000599839.1
ENST00000596362.1 |
CALM3
|
calmodulin 3 (phosphorylase kinase, delta) |
| chr10_-_70092671 | 0.06 |
ENST00000358769.2
ENST00000432941.1 ENST00000495025.2 |
PBLD
|
phenazine biosynthesis-like protein domain containing |
| chr5_-_16936340 | 0.06 |
ENST00000507288.1
ENST00000513610.1 |
MYO10
|
myosin X |
| chrX_+_37639302 | 0.06 |
ENST00000545017.1
ENST00000536160.1 |
CYBB
|
cytochrome b-245, beta polypeptide |
| chr7_+_29234028 | 0.06 |
ENST00000222792.6
|
CHN2
|
chimerin 2 |
| chr17_+_75447326 | 0.06 |
ENST00000591088.1
|
SEPT9
|
septin 9 |
| chr12_+_11081828 | 0.06 |
ENST00000381847.3
ENST00000396400.3 |
PRH2
|
proline-rich protein HaeIII subfamily 2 |
| chr19_+_34287174 | 0.06 |
ENST00000587559.1
ENST00000588637.1 |
KCTD15
|
potassium channel tetramerization domain containing 15 |
| chr12_-_105478339 | 0.06 |
ENST00000424857.2
ENST00000258494.9 |
ALDH1L2
|
aldehyde dehydrogenase 1 family, member L2 |
| chr5_+_140710061 | 0.06 |
ENST00000517417.1
ENST00000378105.3 |
PCDHGA1
|
protocadherin gamma subfamily A, 1 |
| chr16_-_30798492 | 0.05 |
ENST00000262525.4
|
ZNF629
|
zinc finger protein 629 |
| chr8_+_67039278 | 0.05 |
ENST00000276573.7
ENST00000350034.4 |
TRIM55
|
tripartite motif containing 55 |
| chr2_-_161349909 | 0.05 |
ENST00000392753.3
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1 |
| chr21_+_30672433 | 0.05 |
ENST00000451655.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr3_+_107241783 | 0.05 |
ENST00000415149.2
ENST00000402543.1 ENST00000325805.8 ENST00000427402.1 |
BBX
|
bobby sox homolog (Drosophila) |
| chr20_+_48884002 | 0.05 |
ENST00000425497.1
ENST00000445003.1 |
RP11-290F20.3
|
RP11-290F20.3 |
| chr3_+_151986709 | 0.05 |
ENST00000495875.2
ENST00000493459.1 ENST00000324210.5 ENST00000459747.1 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr3_-_62359180 | 0.05 |
ENST00000283268.3
|
FEZF2
|
FEZ family zinc finger 2 |
| chr12_-_109915098 | 0.05 |
ENST00000542858.1
ENST00000542262.1 ENST00000424763.2 |
KCTD10
|
potassium channel tetramerization domain containing 10 |
| chrX_-_10645724 | 0.05 |
ENST00000413894.1
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr6_+_149068464 | 0.05 |
ENST00000367463.4
|
UST
|
uronyl-2-sulfotransferase |
| chr11_-_64013663 | 0.05 |
ENST00000392210.2
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr2_-_160472952 | 0.05 |
ENST00000541068.2
ENST00000355831.2 ENST00000343439.5 ENST00000392782.1 |
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr18_+_42277095 | 0.05 |
ENST00000591940.1
|
SETBP1
|
SET binding protein 1 |
| chr10_-_99094458 | 0.05 |
ENST00000371019.2
|
FRAT2
|
frequently rearranged in advanced T-cell lymphomas 2 |
| chr9_-_134955246 | 0.05 |
ENST00000357028.2
ENST00000474263.1 ENST00000292035.5 |
MED27
|
mediator complex subunit 27 |
| chr11_+_57529234 | 0.05 |
ENST00000360682.6
ENST00000361796.4 ENST00000529526.1 ENST00000426142.2 ENST00000399050.4 ENST00000361391.6 ENST00000361332.4 ENST00000532463.1 ENST00000529986.1 ENST00000358694.6 ENST00000532787.1 ENST00000533667.1 ENST00000532649.1 ENST00000528621.1 ENST00000530748.1 ENST00000428599.2 ENST00000527467.1 ENST00000528232.1 ENST00000531014.1 ENST00000526772.1 ENST00000529873.1 ENST00000525902.1 ENST00000532844.1 ENST00000526357.1 ENST00000530094.1 ENST00000415361.2 ENST00000532245.1 ENST00000534579.1 ENST00000526938.1 |
CTNND1
|
catenin (cadherin-associated protein), delta 1 |
| chr1_+_84767289 | 0.05 |
ENST00000394834.3
ENST00000370669.1 |
SAMD13
|
sterile alpha motif domain containing 13 |
| chr1_+_183774285 | 0.05 |
ENST00000539189.1
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1 |
| chr17_+_19030782 | 0.05 |
ENST00000344415.4
ENST00000577213.1 |
GRAPL
|
GRB2-related adaptor protein-like |
| chr2_-_1748214 | 0.05 |
ENST00000433670.1
ENST00000425171.1 ENST00000252804.4 |
PXDN
|
peroxidasin homolog (Drosophila) |
| chr10_+_114709999 | 0.05 |
ENST00000355995.4
ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr3_-_167813672 | 0.05 |
ENST00000470487.1
|
GOLIM4
|
golgi integral membrane protein 4 |
| chr17_+_75283973 | 0.04 |
ENST00000431235.2
ENST00000449803.2 |
SEPT9
|
septin 9 |
| chr15_+_92937144 | 0.04 |
ENST00000539113.1
ENST00000555434.1 |
ST8SIA2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr10_-_21806759 | 0.04 |
ENST00000444772.3
|
SKIDA1
|
SKI/DACH domain containing 1 |
| chr11_+_126225529 | 0.04 |
ENST00000227495.6
ENST00000444328.2 ENST00000356132.4 |
ST3GAL4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr7_-_99869799 | 0.04 |
ENST00000436886.2
|
GATS
|
GATS, stromal antigen 3 opposite strand |
| chr3_-_48700310 | 0.04 |
ENST00000164024.4
ENST00000544264.1 |
CELSR3
|
cadherin, EGF LAG seven-pass G-type receptor 3 |
| chr15_-_88799948 | 0.04 |
ENST00000394480.2
|
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr12_+_31079652 | 0.04 |
ENST00000546076.1
ENST00000535215.1 ENST00000544427.1 ENST00000261177.9 |
TSPAN11
|
tetraspanin 11 |
| chr5_-_111093081 | 0.04 |
ENST00000453526.2
ENST00000509427.1 |
NREP
|
neuronal regeneration related protein |
| chr10_-_128359074 | 0.04 |
ENST00000544758.1
|
C10orf90
|
chromosome 10 open reading frame 90 |
| chr15_-_88799661 | 0.04 |
ENST00000360948.2
ENST00000357724.2 ENST00000355254.2 ENST00000317501.3 |
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr1_-_50889155 | 0.04 |
ENST00000404795.3
|
DMRTA2
|
DMRT-like family A2 |
| chr7_-_94953878 | 0.04 |
ENST00000222381.3
|
PON1
|
paraoxonase 1 |
| chr2_-_86564696 | 0.04 |
ENST00000437769.1
|
REEP1
|
receptor accessory protein 1 |
| chr3_+_189349162 | 0.04 |
ENST00000264731.3
ENST00000382063.4 ENST00000418709.2 ENST00000320472.5 ENST00000392460.3 ENST00000440651.2 |
TP63
|
tumor protein p63 |
| chr3_-_123339418 | 0.04 |
ENST00000583087.1
|
MYLK
|
myosin light chain kinase |
| chr17_+_7461580 | 0.04 |
ENST00000483039.1
ENST00000396542.1 |
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr20_+_60698180 | 0.04 |
ENST00000361670.3
|
LSM14B
|
LSM14B, SCD6 homolog B (S. cerevisiae) |
| chr19_+_13229126 | 0.04 |
ENST00000292431.4
|
NACC1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr20_-_22559211 | 0.04 |
ENST00000564492.1
|
LINC00261
|
long intergenic non-protein coding RNA 261 |
| chr1_+_228337553 | 0.04 |
ENST00000366714.2
|
GJC2
|
gap junction protein, gamma 2, 47kDa |
| chr7_-_32111009 | 0.04 |
ENST00000396184.3
ENST00000396189.2 ENST00000321453.7 |
PDE1C
|
phosphodiesterase 1C, calmodulin-dependent 70kDa |
| chr17_-_78009647 | 0.04 |
ENST00000310924.2
|
TBC1D16
|
TBC1 domain family, member 16 |
| chr1_-_120190396 | 0.04 |
ENST00000421812.2
|
ZNF697
|
zinc finger protein 697 |
| chr3_-_141747950 | 0.04 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr8_+_67039131 | 0.04 |
ENST00000315962.4
ENST00000353317.5 |
TRIM55
|
tripartite motif containing 55 |
| chr5_+_31193847 | 0.04 |
ENST00000514738.1
ENST00000265071.2 |
CDH6
|
cadherin 6, type 2, K-cadherin (fetal kidney) |
| chr2_-_166060552 | 0.04 |
ENST00000283254.7
ENST00000453007.1 |
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr9_-_130712995 | 0.04 |
ENST00000373084.4
|
FAM102A
|
family with sequence similarity 102, member A |
| chr4_+_150999418 | 0.04 |
ENST00000296550.7
|
DCLK2
|
doublecortin-like kinase 2 |
| chr13_+_78315466 | 0.04 |
ENST00000314070.5
ENST00000462234.1 |
SLAIN1
|
SLAIN motif family, member 1 |
| chr21_+_17792672 | 0.04 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr7_+_134551583 | 0.04 |
ENST00000435928.1
|
CALD1
|
caldesmon 1 |
| chr17_-_49124230 | 0.04 |
ENST00000510283.1
ENST00000510855.1 |
SPAG9
|
sperm associated antigen 9 |
| chr4_-_83351294 | 0.04 |
ENST00000502762.1
|
HNRNPDL
|
heterogeneous nuclear ribonucleoprotein D-like |
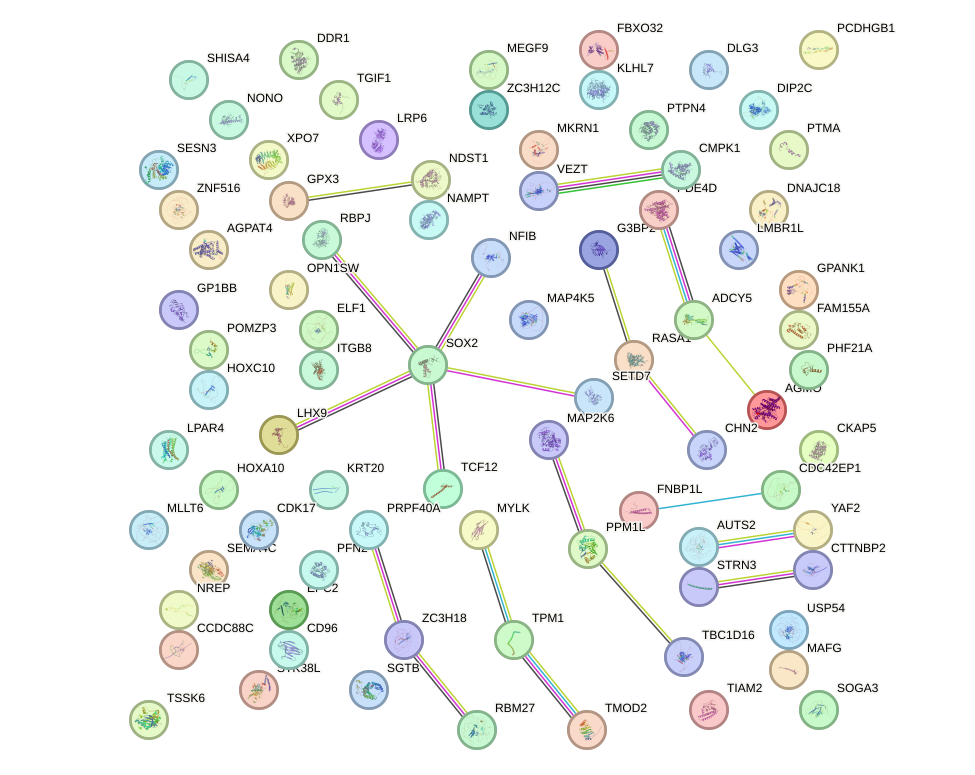
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX10_SOX15
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.1 | 0.2 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.1 | 0.8 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.5 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.1 | GO:0048687 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:0035261 | external genitalia morphogenesis(GO:0035261) |
| 0.0 | 0.2 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.1 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.1 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 | 0.1 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.2 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) myoblast fate commitment(GO:0048625) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.1 | GO:0051466 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.0 | 0.1 | GO:1903450 | regulation of G1 to G0 transition(GO:1903450) positive regulation of G1 to G0 transition(GO:1903452) |
| 0.0 | 0.3 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) |
| 0.0 | 0.3 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.2 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.0 | GO:0034444 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.1 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.1 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 | 0.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.0 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) |
| 0.0 | 0.0 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.0 | 0.1 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.0 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.0 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.2 | GO:1990907 | beta-catenin-TCF7L2 complex(GO:0070369) beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.3 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.1 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.8 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.3 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.3 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.1 | GO:1904928 | coreceptor activity involved in canonical Wnt signaling pathway(GO:1904928) |
| 0.0 | 0.1 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.0 | 0.0 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.0 | 0.1 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.0 | 0.1 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.0 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.0 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.2 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.0 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID ARF 3PATHWAY | Arf1 pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME OPSINS | Genes involved in Opsins |