Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
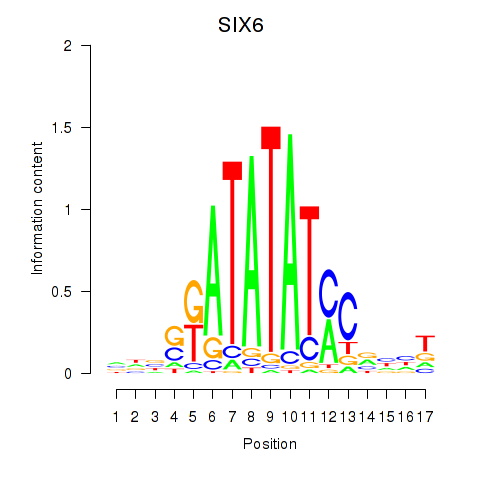
Results for SIX6
Z-value: 0.55
Transcription factors associated with SIX6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SIX6
|
ENSG00000184302.6 | SIX homeobox 6 |
Activity profile of SIX6 motif
Sorted Z-values of SIX6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_9203855 | 0.93 |
ENST00000429566.3
|
OR1M1
|
olfactory receptor, family 1, subfamily M, member 1 |
| chr12_-_71512027 | 0.55 |
ENST00000549357.1
|
CTD-2021H9.3
|
Uncharacterized protein |
| chr6_+_37012607 | 0.33 |
ENST00000423336.1
|
COX6A1P2
|
cytochrome c oxidase subunit VIa polypeptide 1 pseudogene 2 |
| chr4_-_6694189 | 0.30 |
ENST00000596858.1
|
AC093323.1
|
Uncharacterized protein |
| chr6_-_8102279 | 0.28 |
ENST00000488226.2
|
EEF1E1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr9_-_100707116 | 0.27 |
ENST00000259456.3
|
HEMGN
|
hemogen |
| chr12_-_14849470 | 0.21 |
ENST00000261170.3
|
GUCY2C
|
guanylate cyclase 2C (heat stable enterotoxin receptor) |
| chr15_+_63414760 | 0.20 |
ENST00000557972.1
|
LACTB
|
lactamase, beta |
| chr10_+_52094298 | 0.19 |
ENST00000595931.1
|
AC069547.1
|
HCG1745369; PRO3073; Uncharacterized protein |
| chr12_-_121477039 | 0.19 |
ENST00000257570.5
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr12_-_121476959 | 0.19 |
ENST00000339275.5
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr8_+_32579271 | 0.17 |
ENST00000518084.1
|
NRG1
|
neuregulin 1 |
| chr16_+_22524379 | 0.15 |
ENST00000536620.1
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr13_+_73629107 | 0.15 |
ENST00000539231.1
|
KLF5
|
Kruppel-like factor 5 (intestinal) |
| chr12_+_133614062 | 0.15 |
ENST00000540031.1
ENST00000536123.1 |
ZNF84
|
zinc finger protein 84 |
| chr3_+_151531859 | 0.15 |
ENST00000488869.1
|
AADAC
|
arylacetamide deacetylase |
| chr3_-_107596910 | 0.14 |
ENST00000464359.2
ENST00000464823.1 ENST00000466155.1 ENST00000473528.2 ENST00000608306.1 ENST00000488852.1 ENST00000608137.1 ENST00000608307.1 ENST00000609429.1 ENST00000601385.1 ENST00000475362.1 ENST00000600240.1 ENST00000600749.1 |
LINC00635
|
long intergenic non-protein coding RNA 635 |
| chr22_+_24666763 | 0.13 |
ENST00000437398.1
ENST00000421374.1 ENST00000314328.9 ENST00000541492.1 |
SPECC1L
|
sperm antigen with calponin homology and coiled-coil domains 1-like |
| chr12_-_10978957 | 0.12 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr1_+_112016414 | 0.12 |
ENST00000343534.5
ENST00000369718.3 |
C1orf162
|
chromosome 1 open reading frame 162 |
| chr8_+_101170134 | 0.12 |
ENST00000520643.1
|
SPAG1
|
sperm associated antigen 1 |
| chr8_+_32579321 | 0.12 |
ENST00000522402.1
|
NRG1
|
neuregulin 1 |
| chr19_-_14628645 | 0.12 |
ENST00000598235.1
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr1_-_65468148 | 0.11 |
ENST00000415842.1
|
RP11-182I10.3
|
RP11-182I10.3 |
| chr13_+_34392200 | 0.11 |
ENST00000434425.1
|
RFC3
|
replication factor C (activator 1) 3, 38kDa |
| chr7_-_123198284 | 0.11 |
ENST00000355749.2
|
NDUFA5
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 |
| chr12_+_19282643 | 0.11 |
ENST00000317589.4
ENST00000355397.3 ENST00000359180.3 ENST00000309364.4 ENST00000540972.1 ENST00000429027.2 |
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr12_-_46385811 | 0.10 |
ENST00000419565.2
|
SCAF11
|
SR-related CTD-associated factor 11 |
| chr8_+_101170257 | 0.10 |
ENST00000251809.3
|
SPAG1
|
sperm associated antigen 1 |
| chr11_-_104916034 | 0.09 |
ENST00000528513.1
ENST00000375706.2 ENST00000375704.3 |
CARD16
|
caspase recruitment domain family, member 16 |
| chr15_-_80263506 | 0.09 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr12_-_121476750 | 0.09 |
ENST00000543677.1
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr8_+_107593198 | 0.09 |
ENST00000517686.1
|
OXR1
|
oxidation resistance 1 |
| chr1_-_150669604 | 0.08 |
ENST00000427665.1
ENST00000540514.1 |
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr10_-_14614311 | 0.08 |
ENST00000479731.1
ENST00000468492.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr4_+_189321881 | 0.08 |
ENST00000512839.1
ENST00000513313.1 |
LINC01060
|
long intergenic non-protein coding RNA 1060 |
| chr8_+_32579341 | 0.07 |
ENST00000519240.1
ENST00000539990.1 |
NRG1
|
neuregulin 1 |
| chr1_+_179263308 | 0.07 |
ENST00000426956.1
|
SOAT1
|
sterol O-acyltransferase 1 |
| chr3_+_186742464 | 0.07 |
ENST00000416235.1
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr1_+_160709055 | 0.07 |
ENST00000368043.3
ENST00000368042.3 ENST00000458602.2 ENST00000458104.2 |
SLAMF7
|
SLAM family member 7 |
| chr9_-_135230336 | 0.07 |
ENST00000224140.5
ENST00000372169.2 ENST00000393220.1 |
SETX
|
senataxin |
| chr7_+_107332334 | 0.07 |
ENST00000541474.1
ENST00000544569.1 |
SLC26A4
|
solute carrier family 26 (anion exchanger), member 4 |
| chr19_-_16008880 | 0.07 |
ENST00000011989.7
ENST00000221700.6 |
CYP4F2
|
cytochrome P450, family 4, subfamily F, polypeptide 2 |
| chr22_+_18043133 | 0.06 |
ENST00000327451.6
ENST00000399813.1 |
SLC25A18
|
solute carrier family 25 (glutamate carrier), member 18 |
| chr10_-_75008620 | 0.06 |
ENST00000372950.4
|
DNAJC9
|
DnaJ (Hsp40) homolog, subfamily C, member 9 |
| chr4_-_184241927 | 0.06 |
ENST00000323319.5
|
CLDN22
|
claudin 22 |
| chr2_+_177134201 | 0.06 |
ENST00000452865.1
|
MTX2
|
metaxin 2 |
| chr7_+_6121296 | 0.06 |
ENST00000428901.1
|
AC004895.4
|
AC004895.4 |
| chr2_-_100925967 | 0.06 |
ENST00000409647.1
|
LONRF2
|
LON peptidase N-terminal domain and ring finger 2 |
| chr1_-_20446020 | 0.06 |
ENST00000375105.3
|
PLA2G2D
|
phospholipase A2, group IID |
| chr10_+_91061712 | 0.06 |
ENST00000371826.3
|
IFIT2
|
interferon-induced protein with tetratricopeptide repeats 2 |
| chr12_-_15374328 | 0.05 |
ENST00000537647.1
|
RERG
|
RAS-like, estrogen-regulated, growth inhibitor |
| chr6_+_80816342 | 0.05 |
ENST00000369760.4
ENST00000356489.5 ENST00000320393.6 |
BCKDHB
|
branched chain keto acid dehydrogenase E1, beta polypeptide |
| chr17_-_67057047 | 0.05 |
ENST00000495634.1
ENST00000453985.2 ENST00000585714.1 |
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr1_-_36235559 | 0.05 |
ENST00000251195.5
|
CLSPN
|
claspin |
| chr19_+_46000479 | 0.05 |
ENST00000456399.2
|
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr12_-_28123206 | 0.05 |
ENST00000542963.1
ENST00000535992.1 |
PTHLH
|
parathyroid hormone-like hormone |
| chr6_+_127898312 | 0.05 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr17_-_6524159 | 0.05 |
ENST00000589033.1
|
KIAA0753
|
KIAA0753 |
| chr17_-_67224812 | 0.05 |
ENST00000423818.2
|
ABCA10
|
ATP-binding cassette, sub-family A (ABC1), member 10 |
| chr8_+_67104323 | 0.05 |
ENST00000518412.1
ENST00000518035.1 ENST00000517725.1 |
LINC00967
|
long intergenic non-protein coding RNA 967 |
| chr12_-_11091862 | 0.05 |
ENST00000537503.1
|
TAS2R14
|
taste receptor, type 2, member 14 |
| chr15_-_51397473 | 0.05 |
ENST00000327536.5
|
TNFAIP8L3
|
tumor necrosis factor, alpha-induced protein 8-like 3 |
| chr9_-_100684845 | 0.04 |
ENST00000375119.3
|
C9orf156
|
chromosome 9 open reading frame 156 |
| chr1_+_212738676 | 0.04 |
ENST00000366981.4
ENST00000366987.2 |
ATF3
|
activating transcription factor 3 |
| chr2_-_62081148 | 0.04 |
ENST00000404929.1
|
FAM161A
|
family with sequence similarity 161, member A |
| chr4_+_57396766 | 0.04 |
ENST00000512175.2
|
THEGL
|
theg spermatid protein-like |
| chrX_-_99665262 | 0.04 |
ENST00000373034.4
ENST00000255531.7 |
PCDH19
|
protocadherin 19 |
| chr4_+_74301880 | 0.04 |
ENST00000395792.2
ENST00000226359.2 |
AFP
|
alpha-fetoprotein |
| chr17_-_5342380 | 0.04 |
ENST00000225698.4
|
C1QBP
|
complement component 1, q subcomponent binding protein |
| chr7_-_81635106 | 0.04 |
ENST00000443883.1
|
CACNA2D1
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
| chrX_+_128674213 | 0.04 |
ENST00000371113.4
ENST00000357121.5 |
OCRL
|
oculocerebrorenal syndrome of Lowe |
| chr8_+_84824920 | 0.03 |
ENST00000523678.1
|
RP11-120I21.2
|
RP11-120I21.2 |
| chr2_-_100939195 | 0.03 |
ENST00000393437.3
|
LONRF2
|
LON peptidase N-terminal domain and ring finger 2 |
| chr2_+_177134134 | 0.03 |
ENST00000249442.6
ENST00000392529.2 ENST00000443241.1 |
MTX2
|
metaxin 2 |
| chr16_+_72090053 | 0.03 |
ENST00000576168.2
ENST00000567185.3 ENST00000567612.2 |
HP
|
haptoglobin |
| chr10_+_45495898 | 0.03 |
ENST00000298299.3
|
ZNF22
|
zinc finger protein 22 |
| chr20_+_34802295 | 0.03 |
ENST00000432603.1
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr9_-_7800067 | 0.02 |
ENST00000358227.4
|
TMEM261
|
transmembrane protein 261 |
| chrX_-_118699325 | 0.02 |
ENST00000320339.4
ENST00000371594.4 ENST00000536133.1 |
CXorf56
|
chromosome X open reading frame 56 |
| chr18_+_158327 | 0.02 |
ENST00000582707.1
|
USP14
|
ubiquitin specific peptidase 14 (tRNA-guanine transglycosylase) |
| chr22_-_17702729 | 0.02 |
ENST00000449907.2
ENST00000441548.1 ENST00000399839.1 |
CECR1
|
cat eye syndrome chromosome region, candidate 1 |
| chr9_-_99382065 | 0.02 |
ENST00000265659.2
ENST00000375241.1 ENST00000375236.1 |
CDC14B
|
cell division cycle 14B |
| chr13_-_20080080 | 0.02 |
ENST00000400103.2
|
TPTE2
|
transmembrane phosphoinositide 3-phosphatase and tensin homolog 2 |
| chr16_+_19467772 | 0.02 |
ENST00000219821.5
ENST00000561503.1 ENST00000564959.1 |
TMC5
|
transmembrane channel-like 5 |
| chr13_-_46679185 | 0.02 |
ENST00000439329.3
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr5_+_115420688 | 0.02 |
ENST00000274458.4
|
COMMD10
|
COMM domain containing 10 |
| chr9_+_109685630 | 0.02 |
ENST00000451160.2
|
RP11-508N12.4
|
Uncharacterized protein |
| chr13_-_46679144 | 0.02 |
ENST00000181383.4
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chrX_-_20236970 | 0.02 |
ENST00000379548.4
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr1_+_40942887 | 0.02 |
ENST00000372706.1
|
ZFP69
|
ZFP69 zinc finger protein |
| chr16_-_21436459 | 0.02 |
ENST00000448012.2
ENST00000504841.2 ENST00000419180.2 |
NPIPB3
|
nuclear pore complex interacting protein family, member B3 |
| chrX_+_38660704 | 0.02 |
ENST00000378474.3
|
MID1IP1
|
MID1 interacting protein 1 |
| chr11_-_45307817 | 0.02 |
ENST00000020926.3
|
SYT13
|
synaptotagmin XIII |
| chr12_+_117348742 | 0.01 |
ENST00000309909.5
ENST00000455858.2 |
FBXW8
|
F-box and WD repeat domain containing 8 |
| chr1_-_182369751 | 0.01 |
ENST00000367565.1
|
TEDDM1
|
transmembrane epididymal protein 1 |
| chr1_-_67142710 | 0.01 |
ENST00000502413.2
|
AL139147.1
|
Uncharacterized protein |
| chr10_-_79397479 | 0.01 |
ENST00000404771.3
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr12_-_498620 | 0.01 |
ENST00000399788.2
ENST00000382815.4 |
KDM5A
|
lysine (K)-specific demethylase 5A |
| chr3_-_196242233 | 0.01 |
ENST00000397537.2
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr1_+_160709029 | 0.01 |
ENST00000444090.2
ENST00000441662.2 |
SLAMF7
|
SLAM family member 7 |
| chr2_-_58468437 | 0.01 |
ENST00000403676.1
ENST00000427708.2 ENST00000403295.3 ENST00000446381.1 ENST00000417361.1 ENST00000233741.4 ENST00000402135.3 ENST00000540646.1 ENST00000449070.1 |
FANCL
|
Fanconi anemia, complementation group L |
| chr17_+_61271355 | 0.01 |
ENST00000583356.1
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr18_+_42260059 | 0.01 |
ENST00000426838.4
|
SETBP1
|
SET binding protein 1 |
| chr17_+_7788104 | 0.01 |
ENST00000380358.4
|
CHD3
|
chromodomain helicase DNA binding protein 3 |
| chr14_+_21152259 | 0.01 |
ENST00000555835.1
ENST00000336811.6 |
RNASE4
ANG
|
ribonuclease, RNase A family, 4 angiogenin, ribonuclease, RNase A family, 5 |
| chr2_+_85822857 | 0.01 |
ENST00000306368.4
ENST00000414390.1 ENST00000456023.1 |
RNF181
|
ring finger protein 181 |
| chr1_+_160709076 | 0.01 |
ENST00000359331.4
ENST00000495334.1 |
SLAMF7
|
SLAM family member 7 |
| chr1_+_234509186 | 0.00 |
ENST00000366615.4
|
COA6
|
cytochrome c oxidase assembly factor 6 homolog (S. cerevisiae) |
| chr11_-_104972158 | 0.00 |
ENST00000598974.1
ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1
CARD16
CARD17
|
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr4_-_155511887 | 0.00 |
ENST00000302053.3
ENST00000403106.3 |
FGA
|
fibrinogen alpha chain |
| chr5_+_55147205 | 0.00 |
ENST00000396836.2
ENST00000396834.1 ENST00000447346.2 ENST00000359040.5 |
IL31RA
|
interleukin 31 receptor A |
| chr8_+_26247878 | 0.00 |
ENST00000518611.1
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr11_+_126262027 | 0.00 |
ENST00000526311.1
|
ST3GAL4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr1_-_10003372 | 0.00 |
ENST00000377223.1
ENST00000541052.1 ENST00000377213.1 |
LZIC
|
leucine zipper and CTNNBIP1 domain containing |
| chr20_-_4990931 | 0.00 |
ENST00000379333.1
|
SLC23A2
|
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chr1_-_13673511 | 0.00 |
ENST00000344998.3
ENST00000334600.6 |
PRAMEF14
|
PRAME family member 14 |
| chrX_-_53350522 | 0.00 |
ENST00000396435.3
ENST00000375368.5 |
IQSEC2
|
IQ motif and Sec7 domain 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SIX6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.1 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 | 0.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.3 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.9 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:0042361 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.0 | 0.1 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.2 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.0 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.1 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.1 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.1 | GO:0071494 | cellular response to UV-C(GO:0071494) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.0 | 0.0 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.4 | GO:0030673 | axolemma(GO:0030673) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.4 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.0 | 0.1 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.1 | GO:0097258 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.9 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.1 | GO:0003863 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.0 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.0 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.0 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.0 | 0.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |