Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
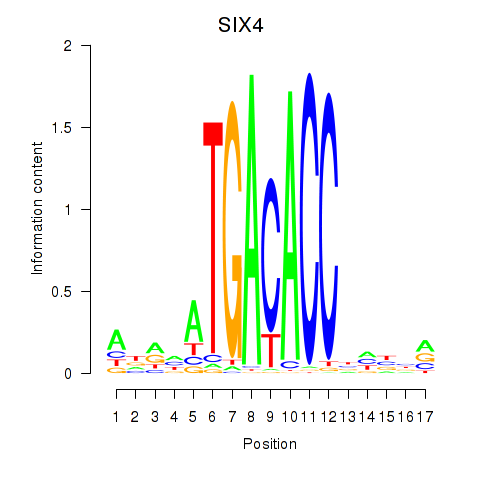
Results for SIX4
Z-value: 0.60
Transcription factors associated with SIX4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SIX4
|
ENSG00000100625.8 | SIX homeobox 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SIX4 | hg19_v2_chr14_-_61190754_61190852 | -0.71 | 2.9e-01 | Click! |
Activity profile of SIX4 motif
Sorted Z-values of SIX4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_191188 | 0.36 |
ENST00000575634.1
|
RPH3AL
|
rabphilin 3A-like (without C2 domains) |
| chr1_+_156611900 | 0.35 |
ENST00000457777.2
ENST00000424639.1 |
BCAN
|
brevican |
| chrX_+_54466829 | 0.30 |
ENST00000375151.4
|
TSR2
|
TSR2, 20S rRNA accumulation, homolog (S. cerevisiae) |
| chr22_-_39268192 | 0.28 |
ENST00000216083.6
|
CBX6
|
chromobox homolog 6 |
| chr6_-_27840099 | 0.26 |
ENST00000328488.2
|
HIST1H3I
|
histone cluster 1, H3i |
| chr19_-_58892389 | 0.24 |
ENST00000427624.2
ENST00000597582.1 |
ZNF837
|
zinc finger protein 837 |
| chr19_+_50270219 | 0.24 |
ENST00000354293.5
ENST00000359032.5 |
AP2A1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr1_+_156611704 | 0.22 |
ENST00000329117.5
|
BCAN
|
brevican |
| chr15_+_41245160 | 0.22 |
ENST00000444189.2
ENST00000446533.3 |
CHAC1
|
ChaC, cation transport regulator homolog 1 (E. coli) |
| chr22_-_30960876 | 0.22 |
ENST00000401975.1
ENST00000428682.1 ENST00000423299.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr3_-_183966717 | 0.22 |
ENST00000446569.1
ENST00000418734.2 ENST00000397676.3 |
ALG3
|
ALG3, alpha-1,3- mannosyltransferase |
| chr22_-_39268308 | 0.22 |
ENST00000407418.3
|
CBX6
|
chromobox homolog 6 |
| chr3_+_183967409 | 0.21 |
ENST00000324557.4
ENST00000402825.3 |
ECE2
|
endothelin converting enzyme 2 |
| chr1_+_156611960 | 0.20 |
ENST00000361588.5
|
BCAN
|
brevican |
| chr19_+_11466062 | 0.19 |
ENST00000251473.5
ENST00000591329.1 ENST00000586380.1 |
DKFZP761J1410
|
Lipid phosphate phosphatase-related protein type 2 |
| chr17_+_7465216 | 0.19 |
ENST00000321337.7
|
SENP3
|
SUMO1/sentrin/SMT3 specific peptidase 3 |
| chr2_-_219134343 | 0.18 |
ENST00000447885.1
ENST00000420660.1 |
AAMP
|
angio-associated, migratory cell protein |
| chr12_-_58135903 | 0.18 |
ENST00000257897.3
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr19_+_50887585 | 0.18 |
ENST00000440232.2
ENST00000601098.1 ENST00000599857.1 ENST00000593887.1 |
POLD1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr12_+_43086018 | 0.17 |
ENST00000550177.1
|
RP11-25I15.3
|
RP11-25I15.3 |
| chr4_-_176828307 | 0.16 |
ENST00000513365.1
ENST00000513667.1 ENST00000503563.1 |
GPM6A
|
glycoprotein M6A |
| chr6_+_160693591 | 0.15 |
ENST00000419196.1
|
RP1-276N6.2
|
RP1-276N6.2 |
| chr19_+_11466167 | 0.15 |
ENST00000591608.1
|
DKFZP761J1410
|
Lipid phosphate phosphatase-related protein type 2 |
| chr2_-_42160486 | 0.14 |
ENST00000427054.1
|
AC104654.2
|
AC104654.2 |
| chr14_+_38065052 | 0.14 |
ENST00000556845.1
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chr12_+_30948600 | 0.14 |
ENST00000550292.1
|
LINC00941
|
long intergenic non-protein coding RNA 941 |
| chr17_-_61523622 | 0.13 |
ENST00000448884.2
ENST00000582297.1 ENST00000582034.1 ENST00000578072.1 ENST00000360793.3 |
CYB561
|
cytochrome b561 |
| chr7_+_2687173 | 0.12 |
ENST00000403167.1
|
TTYH3
|
tweety family member 3 |
| chr1_+_17559776 | 0.11 |
ENST00000537499.1
ENST00000413717.2 ENST00000536552.1 |
PADI1
|
peptidyl arginine deiminase, type I |
| chr3_-_183967296 | 0.11 |
ENST00000455059.1
ENST00000445626.2 |
ALG3
|
ALG3, alpha-1,3- mannosyltransferase |
| chr1_-_154934200 | 0.11 |
ENST00000368457.2
|
PYGO2
|
pygopus family PHD finger 2 |
| chr19_+_13135386 | 0.11 |
ENST00000360105.4
ENST00000588228.1 ENST00000591028.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr2_+_120687335 | 0.11 |
ENST00000544261.1
|
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr19_+_13134772 | 0.11 |
ENST00000587760.1
ENST00000585575.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr11_+_46958248 | 0.11 |
ENST00000536126.1
ENST00000278460.7 ENST00000378618.2 ENST00000395460.2 ENST00000378615.3 ENST00000543718.1 |
C11orf49
|
chromosome 11 open reading frame 49 |
| chr20_+_816695 | 0.11 |
ENST00000246100.3
|
FAM110A
|
family with sequence similarity 110, member A |
| chr4_-_13546632 | 0.10 |
ENST00000382438.5
|
NKX3-2
|
NK3 homeobox 2 |
| chr11_-_46615498 | 0.10 |
ENST00000533727.1
ENST00000534300.1 ENST00000528950.1 ENST00000526606.1 |
AMBRA1
|
autophagy/beclin-1 regulator 1 |
| chr2_-_74710078 | 0.10 |
ENST00000290418.4
|
CCDC142
|
coiled-coil domain containing 142 |
| chr9_+_132427883 | 0.10 |
ENST00000372469.4
|
PRRX2
|
paired related homeobox 2 |
| chr2_+_128458514 | 0.09 |
ENST00000310981.4
|
SFT2D3
|
SFT2 domain containing 3 |
| chr8_+_95907993 | 0.09 |
ENST00000523378.1
|
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr1_+_149239529 | 0.09 |
ENST00000457216.2
|
RP11-403I13.4
|
RP11-403I13.4 |
| chr1_+_15736359 | 0.09 |
ENST00000375980.4
|
EFHD2
|
EF-hand domain family, member D2 |
| chr10_+_102759045 | 0.08 |
ENST00000370220.1
|
LZTS2
|
leucine zipper, putative tumor suppressor 2 |
| chr5_-_1295104 | 0.08 |
ENST00000334602.6
ENST00000508104.2 ENST00000310581.5 ENST00000296820.5 |
TERT
|
telomerase reverse transcriptase |
| chr1_+_8378140 | 0.08 |
ENST00000377479.2
|
SLC45A1
|
solute carrier family 45, member 1 |
| chr2_+_219135115 | 0.08 |
ENST00000248451.3
ENST00000273077.4 |
PNKD
|
paroxysmal nonkinesigenic dyskinesia |
| chr5_+_85913721 | 0.08 |
ENST00000247655.3
ENST00000509578.1 ENST00000515763.1 |
COX7C
|
cytochrome c oxidase subunit VIIc |
| chr7_-_14880892 | 0.08 |
ENST00000406247.3
ENST00000399322.3 ENST00000258767.5 |
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr1_-_207119738 | 0.07 |
ENST00000356495.4
|
PIGR
|
polymeric immunoglobulin receptor |
| chr3_-_49158218 | 0.06 |
ENST00000417901.1
ENST00000306026.5 ENST00000434032.2 |
USP19
|
ubiquitin specific peptidase 19 |
| chr1_-_32264356 | 0.06 |
ENST00000452755.2
|
SPOCD1
|
SPOC domain containing 1 |
| chr11_+_2415061 | 0.06 |
ENST00000481687.1
|
CD81
|
CD81 molecule |
| chr3_-_69249863 | 0.06 |
ENST00000478263.1
ENST00000462512.1 |
FRMD4B
|
FERM domain containing 4B |
| chr3_-_93692681 | 0.06 |
ENST00000348974.4
|
PROS1
|
protein S (alpha) |
| chr11_-_96076334 | 0.05 |
ENST00000524717.1
|
MAML2
|
mastermind-like 2 (Drosophila) |
| chr12_-_10282742 | 0.05 |
ENST00000298523.5
ENST00000396484.2 ENST00000310002.4 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chrX_-_110655391 | 0.05 |
ENST00000356915.2
ENST00000356220.3 |
DCX
|
doublecortin |
| chr7_+_69064300 | 0.05 |
ENST00000342771.4
|
AUTS2
|
autism susceptibility candidate 2 |
| chr20_-_43150601 | 0.05 |
ENST00000541235.1
ENST00000255175.1 ENST00000342374.4 |
SERINC3
|
serine incorporator 3 |
| chrY_+_22737678 | 0.04 |
ENST00000382772.3
|
EIF1AY
|
eukaryotic translation initiation factor 1A, Y-linked |
| chr9_-_95298314 | 0.04 |
ENST00000344604.5
ENST00000375540.1 |
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr11_+_123325106 | 0.04 |
ENST00000525757.1
|
LINC01059
|
long intergenic non-protein coding RNA 1059 |
| chr7_-_26578407 | 0.04 |
ENST00000242109.3
|
KIAA0087
|
KIAA0087 |
| chr5_+_78365536 | 0.04 |
ENST00000255192.3
|
BHMT2
|
betaine--homocysteine S-methyltransferase 2 |
| chr4_-_2420357 | 0.03 |
ENST00000511071.1
ENST00000509171.1 ENST00000290974.2 |
ZFYVE28
|
zinc finger, FYVE domain containing 28 |
| chr2_+_74710194 | 0.03 |
ENST00000410003.1
ENST00000442235.2 ENST00000233623.5 |
TTC31
|
tetratricopeptide repeat domain 31 |
| chr1_+_38022572 | 0.03 |
ENST00000541606.1
|
DNALI1
|
dynein, axonemal, light intermediate chain 1 |
| chr12_+_133757995 | 0.03 |
ENST00000536435.2
ENST00000228289.5 ENST00000541211.2 ENST00000500625.3 ENST00000539248.2 ENST00000542711.2 ENST00000536899.2 ENST00000542986.2 ENST00000537565.1 ENST00000541975.2 |
ZNF268
|
zinc finger protein 268 |
| chr6_+_41040678 | 0.03 |
ENST00000341376.6
ENST00000353205.5 |
NFYA
|
nuclear transcription factor Y, alpha |
| chr1_-_173176452 | 0.03 |
ENST00000281834.3
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4 |
| chr2_-_113999260 | 0.03 |
ENST00000468980.2
|
PAX8
|
paired box 8 |
| chr19_+_19174795 | 0.03 |
ENST00000318596.7
|
SLC25A42
|
solute carrier family 25, member 42 |
| chr7_-_111428957 | 0.03 |
ENST00000417165.1
|
DOCK4
|
dedicator of cytokinesis 4 |
| chr11_+_117049445 | 0.02 |
ENST00000324225.4
ENST00000532960.1 |
SIDT2
|
SID1 transmembrane family, member 2 |
| chrX_-_19504642 | 0.02 |
ENST00000469203.2
|
MAP3K15
|
mitogen-activated protein kinase kinase kinase 15 |
| chr19_+_13135439 | 0.02 |
ENST00000586873.1
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr1_+_116184566 | 0.02 |
ENST00000355485.2
ENST00000369510.4 |
VANGL1
|
VANGL planar cell polarity protein 1 |
| chr5_-_78365437 | 0.02 |
ENST00000380311.4
ENST00000540686.1 ENST00000255189.3 |
DMGDH
|
dimethylglycine dehydrogenase |
| chr17_-_72527605 | 0.02 |
ENST00000392621.1
ENST00000314401.3 |
CD300LB
|
CD300 molecule-like family member b |
| chr10_+_89622870 | 0.01 |
ENST00000371953.3
|
PTEN
|
phosphatase and tensin homolog |
| chrX_+_135388147 | 0.01 |
ENST00000394141.1
|
GPR112
|
G protein-coupled receptor 112 |
| chr9_-_95298254 | 0.01 |
ENST00000444490.2
|
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr12_+_133758115 | 0.01 |
ENST00000541009.2
ENST00000592241.1 |
ZNF268
|
zinc finger protein 268 |
| chr7_-_38969150 | 0.01 |
ENST00000418457.2
|
VPS41
|
vacuolar protein sorting 41 homolog (S. cerevisiae) |
| chr1_-_114414316 | 0.01 |
ENST00000528414.1
ENST00000538253.1 ENST00000460620.1 ENST00000420377.2 ENST00000525799.1 ENST00000359785.5 |
PTPN22
|
protein tyrosine phosphatase, non-receptor type 22 (lymphoid) |
| chr4_-_2420335 | 0.01 |
ENST00000503000.1
|
ZFYVE28
|
zinc finger, FYVE domain containing 28 |
| chr8_+_56014949 | 0.01 |
ENST00000327381.6
|
XKR4
|
XK, Kell blood group complex subunit-related family, member 4 |
| chr1_+_231473743 | 0.00 |
ENST00000295050.7
|
SPRTN
|
SprT-like N-terminal domain |
| chr2_-_219134822 | 0.00 |
ENST00000444053.1
ENST00000248450.4 |
AAMP
|
angio-associated, migratory cell protein |
| chrX_+_70503037 | 0.00 |
ENST00000535149.1
|
NONO
|
non-POU domain containing, octamer-binding |
| chr11_+_117049910 | 0.00 |
ENST00000431081.2
ENST00000524842.1 |
SIDT2
|
SID1 transmembrane family, member 2 |
| chr12_-_10282836 | 0.00 |
ENST00000304084.8
ENST00000353231.5 ENST00000525605.1 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr11_+_117049854 | 0.00 |
ENST00000278951.7
|
SIDT2
|
SID1 transmembrane family, member 2 |
| chr3_-_87325612 | 0.00 |
ENST00000561167.1
ENST00000560656.1 ENST00000344265.3 |
POU1F1
|
POU class 1 homeobox 1 |
| chrX_+_41583408 | 0.00 |
ENST00000302548.4
|
GPR82
|
G protein-coupled receptor 82 |
| chr1_-_152196669 | 0.00 |
ENST00000368801.2
|
HRNR
|
hornerin |
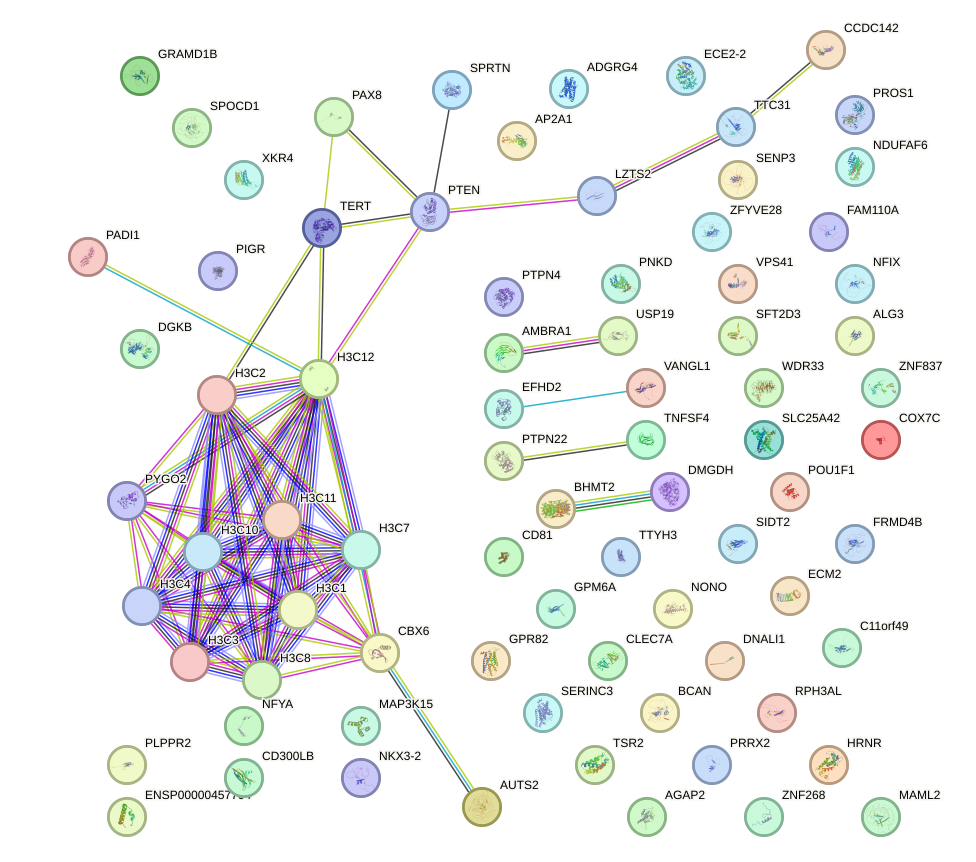
Network of associatons between targets according to the STRING database.
First level regulatory network of SIX4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.8 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.2 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.1 | GO:1900369 | transcription, RNA-templated(GO:0001172) negative regulation of RNA interference(GO:1900369) |
| 0.0 | 0.1 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.2 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.0 | 0.0 | GO:0046022 | positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.3 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.2 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 0.1 | GO:0043128 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0031379 | RNA-directed RNA polymerase complex(GO:0031379) |
| 0.0 | 0.2 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 0.2 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0035034 | histone acetyltransferase regulator activity(GO:0035034) |
| 0.0 | 0.1 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.0 | 0.1 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.2 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.2 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.3 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |