Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
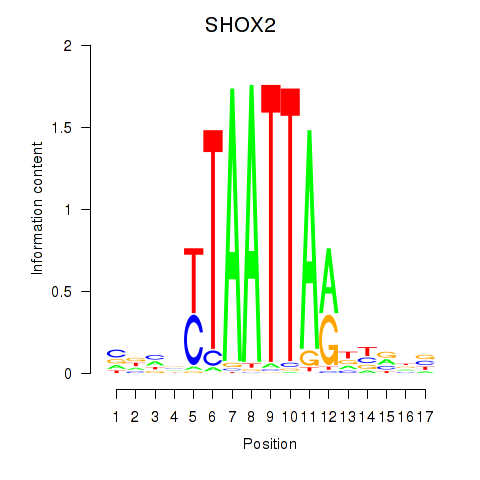
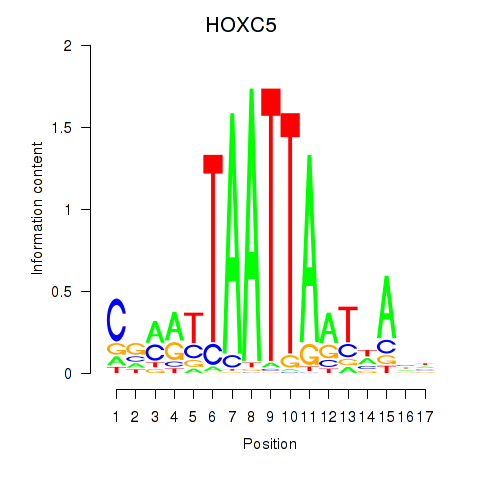
Results for SHOX2_HOXC5
Z-value: 0.18
Transcription factors associated with SHOX2_HOXC5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SHOX2
|
ENSG00000168779.15 | short stature homeobox 2 |
|
HOXC5
|
ENSG00000172789.3 | homeobox C5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXC5 | hg19_v2_chr12_+_54426637_54426637 | -1.00 | 3.4e-03 | Click! |
| SHOX2 | hg19_v2_chr3_-_157823839_157824078 | -0.85 | 1.5e-01 | Click! |
Activity profile of SHOX2_HOXC5 motif
Sorted Z-values of SHOX2_HOXC5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_25150373 | 0.16 |
ENST00000549828.1
|
C12orf77
|
chromosome 12 open reading frame 77 |
| chr6_+_24126350 | 0.15 |
ENST00000378491.4
ENST00000378478.1 ENST00000378477.2 |
NRSN1
|
neurensin 1 |
| chr6_-_26235206 | 0.14 |
ENST00000244534.5
|
HIST1H1D
|
histone cluster 1, H1d |
| chr6_+_26104104 | 0.10 |
ENST00000377803.2
|
HIST1H4C
|
histone cluster 1, H4c |
| chr22_-_32766972 | 0.09 |
ENST00000382084.4
ENST00000382086.2 |
RFPL3S
|
RFPL3 antisense |
| chr5_+_42756903 | 0.09 |
ENST00000361970.5
ENST00000388827.4 |
CCDC152
|
coiled-coil domain containing 152 |
| chr13_-_30160925 | 0.08 |
ENST00000450494.1
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr11_+_29181503 | 0.07 |
ENST00000530960.1
|
RP11-466I1.1
|
RP11-466I1.1 |
| chr12_+_78359999 | 0.07 |
ENST00000550503.1
|
NAV3
|
neuron navigator 3 |
| chr22_-_32767017 | 0.07 |
ENST00000400234.1
|
RFPL3S
|
RFPL3 antisense |
| chr5_+_129083772 | 0.07 |
ENST00000564719.1
|
KIAA1024L
|
KIAA1024-like |
| chr6_-_27835357 | 0.06 |
ENST00000331442.3
|
HIST1H1B
|
histone cluster 1, H1b |
| chr17_+_19091325 | 0.06 |
ENST00000584923.1
|
SNORD3A
|
small nucleolar RNA, C/D box 3A |
| chr2_+_90198535 | 0.06 |
ENST00000390276.2
|
IGKV1D-12
|
immunoglobulin kappa variable 1D-12 |
| chr2_+_232316906 | 0.06 |
ENST00000370380.2
|
AC017104.2
|
Uncharacterized protein |
| chr19_-_58864848 | 0.05 |
ENST00000263100.3
|
A1BG
|
alpha-1-B glycoprotein |
| chr8_+_101349823 | 0.05 |
ENST00000519566.1
|
KB-1991G8.1
|
KB-1991G8.1 |
| chr8_+_39972170 | 0.05 |
ENST00000521257.1
|
RP11-359E19.2
|
RP11-359E19.2 |
| chr3_-_167191814 | 0.05 |
ENST00000466903.1
ENST00000264677.4 |
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2 |
| chr6_-_116833500 | 0.05 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr6_+_26045603 | 0.05 |
ENST00000540144.1
|
HIST1H3C
|
histone cluster 1, H3c |
| chr2_+_182850551 | 0.05 |
ENST00000452904.1
ENST00000409137.3 ENST00000280295.3 |
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr17_-_19015945 | 0.05 |
ENST00000573866.2
|
SNORD3D
|
small nucleolar RNA, C/D box 3D |
| chr5_+_53686658 | 0.05 |
ENST00000512618.1
|
LINC01033
|
long intergenic non-protein coding RNA 1033 |
| chr16_+_15489603 | 0.05 |
ENST00000568766.1
ENST00000287594.7 |
RP11-1021N1.1
MPV17L
|
Uncharacterized protein MPV17 mitochondrial membrane protein-like |
| chr3_+_28390637 | 0.05 |
ENST00000420223.1
ENST00000383768.2 |
ZCWPW2
|
zinc finger, CW type with PWWP domain 2 |
| chr6_-_26032288 | 0.04 |
ENST00000244661.2
|
HIST1H3B
|
histone cluster 1, H3b |
| chr8_-_38008783 | 0.04 |
ENST00000276449.4
|
STAR
|
steroidogenic acute regulatory protein |
| chr7_-_99716914 | 0.04 |
ENST00000431404.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr15_-_54267147 | 0.04 |
ENST00000558866.1
ENST00000558920.1 |
RP11-643A5.2
|
RP11-643A5.2 |
| chr2_-_236684438 | 0.04 |
ENST00000409661.1
|
AC064874.1
|
Uncharacterized protein |
| chr6_+_26183958 | 0.04 |
ENST00000356530.3
|
HIST1H2BE
|
histone cluster 1, H2be |
| chr1_-_201140673 | 0.04 |
ENST00000367333.2
|
TMEM9
|
transmembrane protein 9 |
| chr20_-_29896388 | 0.04 |
ENST00000400549.1
|
DEFB116
|
defensin, beta 116 |
| chr3_-_12587055 | 0.04 |
ENST00000564146.3
|
C3orf83
|
chromosome 3 open reading frame 83 |
| chr3_+_51851612 | 0.04 |
ENST00000456080.1
|
IQCF3
|
IQ motif containing F3 |
| chr15_-_36544450 | 0.04 |
ENST00000561394.1
|
RP11-184D12.1
|
RP11-184D12.1 |
| chr6_+_26156551 | 0.04 |
ENST00000304218.3
|
HIST1H1E
|
histone cluster 1, H1e |
| chrX_-_55208866 | 0.04 |
ENST00000545075.1
|
MTRNR2L10
|
MT-RNR2-like 10 |
| chr21_-_19858196 | 0.04 |
ENST00000422787.1
|
TMPRSS15
|
transmembrane protease, serine 15 |
| chr4_+_9446156 | 0.04 |
ENST00000334879.1
|
DEFB131
|
defensin, beta 131 |
| chr1_+_81001398 | 0.04 |
ENST00000418041.1
ENST00000443104.1 |
RP5-887A10.1
|
RP5-887A10.1 |
| chr16_+_15489629 | 0.03 |
ENST00000396385.3
|
MPV17L
|
MPV17 mitochondrial membrane protein-like |
| chr12_-_11214893 | 0.03 |
ENST00000533467.1
|
TAS2R46
|
taste receptor, type 2, member 46 |
| chr10_-_13043697 | 0.03 |
ENST00000378825.3
|
CCDC3
|
coiled-coil domain containing 3 |
| chr3_+_38537960 | 0.03 |
ENST00000453767.1
|
EXOG
|
endo/exonuclease (5'-3'), endonuclease G-like |
| chr11_+_122733011 | 0.03 |
ENST00000533709.1
|
CRTAM
|
cytotoxic and regulatory T cell molecule |
| chr17_-_4938712 | 0.03 |
ENST00000254853.5
ENST00000424747.1 |
SLC52A1
|
solute carrier family 52 (riboflavin transporter), member 1 |
| chr19_+_52873166 | 0.03 |
ENST00000424032.2
ENST00000600321.1 ENST00000344085.5 ENST00000597976.1 ENST00000422689.2 |
ZNF880
|
zinc finger protein 880 |
| chr6_+_26365443 | 0.03 |
ENST00000527422.1
ENST00000356386.2 ENST00000396934.3 ENST00000377708.2 ENST00000396948.1 ENST00000508906.2 |
BTN3A2
|
butyrophilin, subfamily 3, member A2 |
| chr4_-_112993808 | 0.03 |
ENST00000511219.1
|
RP11-269F21.3
|
RP11-269F21.3 |
| chr22_+_21996549 | 0.03 |
ENST00000248958.4
|
SDF2L1
|
stromal cell-derived factor 2-like 1 |
| chr7_-_111032971 | 0.03 |
ENST00000450877.1
|
IMMP2L
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr22_+_23487513 | 0.03 |
ENST00000263116.2
ENST00000341989.4 |
RAB36
|
RAB36, member RAS oncogene family |
| chr1_+_161494036 | 0.03 |
ENST00000309758.4
|
HSPA6
|
heat shock 70kDa protein 6 (HSP70B') |
| chr12_+_8849773 | 0.03 |
ENST00000541044.1
|
RIMKLB
|
ribosomal modification protein rimK-like family member B |
| chr12_-_70093235 | 0.03 |
ENST00000266661.4
|
BEST3
|
bestrophin 3 |
| chr8_+_62200509 | 0.03 |
ENST00000519846.1
ENST00000518592.1 ENST00000325897.4 |
CLVS1
|
clavesin 1 |
| chr17_+_7155819 | 0.03 |
ENST00000570322.1
ENST00000576496.1 ENST00000574841.2 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr12_-_118628315 | 0.03 |
ENST00000540561.1
|
TAOK3
|
TAO kinase 3 |
| chr14_-_67878917 | 0.03 |
ENST00000216446.4
|
PLEK2
|
pleckstrin 2 |
| chr6_-_26285737 | 0.03 |
ENST00000377727.1
ENST00000289352.1 |
HIST1H4H
|
histone cluster 1, H4h |
| chr3_-_20053741 | 0.03 |
ENST00000389050.4
|
PP2D1
|
protein phosphatase 2C-like domain containing 1 |
| chr3_+_52245458 | 0.03 |
ENST00000459884.1
|
ALAS1
|
aminolevulinate, delta-, synthase 1 |
| chr14_+_61449197 | 0.03 |
ENST00000533744.2
|
SLC38A6
|
solute carrier family 38, member 6 |
| chr19_-_2944907 | 0.03 |
ENST00000314531.4
|
ZNF77
|
zinc finger protein 77 |
| chr5_+_150639360 | 0.03 |
ENST00000523004.1
|
GM2A
|
GM2 ganglioside activator |
| chr19_-_53758094 | 0.03 |
ENST00000601828.1
ENST00000598513.1 ENST00000599012.1 ENST00000333952.4 ENST00000598806.1 |
ZNF677
|
zinc finger protein 677 |
| chr2_+_74648848 | 0.03 |
ENST00000409791.1
ENST00000426787.1 ENST00000348227.4 |
WDR54
|
WD repeat domain 54 |
| chr6_-_27860956 | 0.03 |
ENST00000359611.2
|
HIST1H2AM
|
histone cluster 1, H2am |
| chr15_+_62853562 | 0.03 |
ENST00000561311.1
|
TLN2
|
talin 2 |
| chr6_+_4087664 | 0.03 |
ENST00000430835.2
|
C6orf201
|
chromosome 6 open reading frame 201 |
| chr2_-_114300213 | 0.03 |
ENST00000446595.1
ENST00000416105.1 ENST00000450636.1 ENST00000416758.1 |
RP11-395L14.4
|
RP11-395L14.4 |
| chr1_+_28261533 | 0.03 |
ENST00000411604.1
ENST00000373888.4 |
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr12_+_128399965 | 0.03 |
ENST00000540882.1
ENST00000542089.1 |
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chr7_+_115862858 | 0.03 |
ENST00000393481.2
|
TES
|
testis derived transcript (3 LIM domains) |
| chr13_-_76111945 | 0.02 |
ENST00000355801.4
ENST00000406936.3 |
COMMD6
|
COMM domain containing 6 |
| chr15_+_75970672 | 0.02 |
ENST00000435356.1
|
AC105020.1
|
Uncharacterized protein; cDNA FLJ12988 fis, clone NT2RP3000080 |
| chr8_+_104310661 | 0.02 |
ENST00000522566.1
|
FZD6
|
frizzled family receptor 6 |
| chr15_+_78830023 | 0.02 |
ENST00000599596.1
|
AC027228.1
|
HCG2005369; Uncharacterized protein |
| chr12_-_25348007 | 0.02 |
ENST00000354189.5
ENST00000545133.1 ENST00000554347.1 ENST00000395987.3 ENST00000320267.9 ENST00000395990.2 ENST00000537577.1 |
CASC1
|
cancer susceptibility candidate 1 |
| chr20_-_44600810 | 0.02 |
ENST00000322927.2
ENST00000426788.1 |
ZNF335
|
zinc finger protein 335 |
| chr11_+_92085262 | 0.02 |
ENST00000298047.6
ENST00000409404.2 ENST00000541502.1 |
FAT3
|
FAT atypical cadherin 3 |
| chr1_-_45140074 | 0.02 |
ENST00000420706.1
ENST00000372235.3 ENST00000372242.3 ENST00000372243.3 ENST00000372244.3 |
TMEM53
|
transmembrane protein 53 |
| chrX_+_37639302 | 0.02 |
ENST00000545017.1
ENST00000536160.1 |
CYBB
|
cytochrome b-245, beta polypeptide |
| chr19_-_10227503 | 0.02 |
ENST00000593054.1
|
EIF3G
|
eukaryotic translation initiation factor 3, subunit G |
| chr15_-_75248954 | 0.02 |
ENST00000499788.2
|
RPP25
|
ribonuclease P/MRP 25kDa subunit |
| chr12_-_70093111 | 0.02 |
ENST00000548658.1
ENST00000476098.1 ENST00000331471.4 ENST00000393365.1 |
BEST3
|
bestrophin 3 |
| chr12_-_10601963 | 0.02 |
ENST00000543893.1
|
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr15_+_75639372 | 0.02 |
ENST00000566313.1
ENST00000568059.1 ENST00000568881.1 |
NEIL1
|
nei endonuclease VIII-like 1 (E. coli) |
| chr11_-_124981475 | 0.02 |
ENST00000532156.1
ENST00000532407.1 ENST00000279968.4 ENST00000527766.1 ENST00000529583.1 ENST00000524373.1 ENST00000527271.1 ENST00000526175.1 ENST00000529609.1 ENST00000533273.1 ENST00000531909.1 ENST00000529530.1 |
TMEM218
|
transmembrane protein 218 |
| chr20_-_43133491 | 0.02 |
ENST00000411544.1
|
SERINC3
|
serine incorporator 3 |
| chr19_+_45417921 | 0.02 |
ENST00000252491.4
ENST00000592885.1 ENST00000589781.1 |
APOC1
|
apolipoprotein C-I |
| chr7_-_144533074 | 0.02 |
ENST00000360057.3
ENST00000378099.3 |
TPK1
|
thiamin pyrophosphokinase 1 |
| chr5_+_140227357 | 0.02 |
ENST00000378122.3
|
PCDHA9
|
protocadherin alpha 9 |
| chr19_-_13227514 | 0.02 |
ENST00000587487.1
ENST00000592814.1 |
TRMT1
|
tRNA methyltransferase 1 homolog (S. cerevisiae) |
| chr10_-_28571015 | 0.02 |
ENST00000375719.3
ENST00000375732.1 |
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr1_-_6420737 | 0.02 |
ENST00000541130.1
ENST00000377845.3 |
ACOT7
|
acyl-CoA thioesterase 7 |
| chr19_-_13227463 | 0.02 |
ENST00000437766.1
ENST00000221504.8 |
TRMT1
|
tRNA methyltransferase 1 homolog (S. cerevisiae) |
| chr1_+_43291220 | 0.02 |
ENST00000372514.3
|
ERMAP
|
erythroblast membrane-associated protein (Scianna blood group) |
| chrX_+_72783026 | 0.02 |
ENST00000373504.6
ENST00000373502.5 |
CHIC1
|
cysteine-rich hydrophobic domain 1 |
| chr19_+_50016411 | 0.02 |
ENST00000426395.3
ENST00000600273.1 ENST00000599988.1 |
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr16_-_66764119 | 0.02 |
ENST00000569320.1
|
DYNC1LI2
|
dynein, cytoplasmic 1, light intermediate chain 2 |
| chr7_-_37024665 | 0.02 |
ENST00000396040.2
|
ELMO1
|
engulfment and cell motility 1 |
| chr17_+_39421591 | 0.02 |
ENST00000391355.1
|
KRTAP9-6
|
keratin associated protein 9-6 |
| chr6_+_161123270 | 0.02 |
ENST00000366924.2
ENST00000308192.9 ENST00000418964.1 |
PLG
|
plasminogen |
| chr6_+_27861190 | 0.02 |
ENST00000303806.4
|
HIST1H2BO
|
histone cluster 1, H2bo |
| chr13_+_78315295 | 0.02 |
ENST00000351546.3
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr1_+_16083154 | 0.02 |
ENST00000375771.1
|
FBLIM1
|
filamin binding LIM protein 1 |
| chr16_-_28634874 | 0.02 |
ENST00000395609.1
ENST00000350842.4 |
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr3_-_46000146 | 0.02 |
ENST00000438446.1
|
FYCO1
|
FYVE and coiled-coil domain containing 1 |
| chr11_+_115498761 | 0.02 |
ENST00000424313.2
|
AP000997.1
|
AP000997.1 |
| chr8_+_58890917 | 0.02 |
ENST00000522992.1
|
RP11-1112C15.1
|
RP11-1112C15.1 |
| chr7_+_102553430 | 0.02 |
ENST00000339431.4
ENST00000249377.4 |
LRRC17
|
leucine rich repeat containing 17 |
| chr21_-_27423339 | 0.02 |
ENST00000415997.1
|
APP
|
amyloid beta (A4) precursor protein |
| chr3_+_187871659 | 0.02 |
ENST00000416784.1
ENST00000430340.1 ENST00000414139.1 ENST00000454789.1 |
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr18_-_59274139 | 0.02 |
ENST00000586949.1
|
RP11-879F14.2
|
RP11-879F14.2 |
| chr14_+_56584414 | 0.02 |
ENST00000559044.1
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr5_+_81575281 | 0.02 |
ENST00000380167.4
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like |
| chr8_-_66474884 | 0.02 |
ENST00000520902.1
|
CTD-3025N20.2
|
CTD-3025N20.2 |
| chr1_+_28261492 | 0.02 |
ENST00000373894.3
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chrX_+_47444613 | 0.02 |
ENST00000445623.1
|
TIMP1
|
TIMP metallopeptidase inhibitor 1 |
| chr14_-_81425828 | 0.02 |
ENST00000555529.1
ENST00000556042.1 ENST00000556981.1 |
CEP128
|
centrosomal protein 128kDa |
| chr11_+_119205222 | 0.02 |
ENST00000311413.4
|
RNF26
|
ring finger protein 26 |
| chr6_+_160693591 | 0.02 |
ENST00000419196.1
|
RP1-276N6.2
|
RP1-276N6.2 |
| chr19_-_56826157 | 0.02 |
ENST00000592509.1
ENST00000592679.1 ENST00000588442.1 ENST00000593106.1 ENST00000587492.1 ENST00000254165.3 |
ZSCAN5A
|
zinc finger and SCAN domain containing 5A |
| chr4_-_120243545 | 0.02 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr10_-_128110441 | 0.02 |
ENST00000456514.1
|
LINC00601
|
long intergenic non-protein coding RNA 601 |
| chr18_+_616672 | 0.02 |
ENST00000338387.7
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr11_-_62609281 | 0.02 |
ENST00000525239.1
ENST00000538098.2 |
WDR74
|
WD repeat domain 74 |
| chr5_-_149829244 | 0.02 |
ENST00000312037.5
|
RPS14
|
ribosomal protein S14 |
| chr11_+_114168085 | 0.02 |
ENST00000541754.1
|
NNMT
|
nicotinamide N-methyltransferase |
| chr6_-_26216872 | 0.02 |
ENST00000244601.3
|
HIST1H2BG
|
histone cluster 1, H2bg |
| chr19_+_50016610 | 0.02 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr14_+_61449076 | 0.02 |
ENST00000526105.1
|
SLC38A6
|
solute carrier family 38, member 6 |
| chr8_-_41166953 | 0.02 |
ENST00000220772.3
|
SFRP1
|
secreted frizzled-related protein 1 |
| chr5_+_140529630 | 0.02 |
ENST00000543635.1
|
PCDHB6
|
protocadherin beta 6 |
| chr6_+_63921351 | 0.02 |
ENST00000370659.1
|
FKBP1C
|
FK506 binding protein 1C |
| chr2_-_86333244 | 0.02 |
ENST00000263857.6
ENST00000409681.1 |
POLR1A
|
polymerase (RNA) I polypeptide A, 194kDa |
| chr6_+_52442083 | 0.02 |
ENST00000606714.1
|
TRAM2-AS1
|
TRAM2 antisense RNA 1 (head to head) |
| chr22_+_50981079 | 0.02 |
ENST00000609268.1
|
CTA-384D8.34
|
CTA-384D8.34 |
| chr11_-_58611957 | 0.02 |
ENST00000532258.1
|
GLYATL2
|
glycine-N-acyltransferase-like 2 |
| chr19_-_51920952 | 0.02 |
ENST00000356298.5
ENST00000339313.5 ENST00000529627.1 ENST00000439889.2 ENST00000353836.5 ENST00000432469.2 |
SIGLEC10
|
sialic acid binding Ig-like lectin 10 |
| chr19_-_13227534 | 0.02 |
ENST00000588229.1
ENST00000357720.4 |
TRMT1
|
tRNA methyltransferase 1 homolog (S. cerevisiae) |
| chrX_-_92928557 | 0.02 |
ENST00000373079.3
ENST00000475430.2 |
NAP1L3
|
nucleosome assembly protein 1-like 3 |
| chr10_-_74283694 | 0.02 |
ENST00000398763.4
ENST00000418483.2 ENST00000489666.2 |
MICU1
|
mitochondrial calcium uptake 1 |
| chr12_-_59314246 | 0.02 |
ENST00000320743.3
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3 |
| chr8_-_90769422 | 0.02 |
ENST00000524190.1
ENST00000523859.1 |
RP11-37B2.1
|
RP11-37B2.1 |
| chr7_-_102184083 | 0.02 |
ENST00000379357.5
|
POLR2J3
|
polymerase (RNA) II (DNA directed) polypeptide J3 |
| chr14_+_35591928 | 0.02 |
ENST00000605870.1
ENST00000557404.3 |
KIAA0391
|
KIAA0391 |
| chr6_+_26204825 | 0.02 |
ENST00000360441.4
|
HIST1H4E
|
histone cluster 1, H4e |
| chr1_-_114414316 | 0.02 |
ENST00000528414.1
ENST00000538253.1 ENST00000460620.1 ENST00000420377.2 ENST00000525799.1 ENST00000359785.5 |
PTPN22
|
protein tyrosine phosphatase, non-receptor type 22 (lymphoid) |
| chr5_-_139726181 | 0.02 |
ENST00000507104.1
ENST00000230990.6 |
HBEGF
|
heparin-binding EGF-like growth factor |
| chr14_-_54423529 | 0.02 |
ENST00000245451.4
ENST00000559087.1 |
BMP4
|
bone morphogenetic protein 4 |
| chr6_-_32145861 | 0.02 |
ENST00000336984.6
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr1_+_196743912 | 0.02 |
ENST00000367425.4
|
CFHR3
|
complement factor H-related 3 |
| chr17_-_40828969 | 0.02 |
ENST00000591022.1
ENST00000587627.1 ENST00000293349.6 |
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr14_-_35591156 | 0.02 |
ENST00000554361.1
|
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma |
| chrX_+_22050546 | 0.02 |
ENST00000379374.4
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked |
| chr19_+_41284121 | 0.02 |
ENST00000594800.1
ENST00000357052.2 ENST00000602173.1 |
RAB4B
|
RAB4B, member RAS oncogene family |
| chr14_-_23426231 | 0.02 |
ENST00000556915.1
|
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr17_+_79650962 | 0.02 |
ENST00000329138.4
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate |
| chr1_+_81106951 | 0.02 |
ENST00000443565.1
|
RP5-887A10.1
|
RP5-887A10.1 |
| chr14_+_100532771 | 0.02 |
ENST00000557153.1
|
EVL
|
Enah/Vasp-like |
| chr4_-_113558079 | 0.02 |
ENST00000445203.2
|
C4orf21
|
chromosome 4 open reading frame 21 |
| chr6_+_43395272 | 0.02 |
ENST00000372530.4
|
ABCC10
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 10 |
| chr12_+_128399917 | 0.02 |
ENST00000544645.1
|
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chr2_-_225811747 | 0.02 |
ENST00000409592.3
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr2_-_112237835 | 0.02 |
ENST00000442293.1
ENST00000439494.1 |
MIR4435-1HG
|
MIR4435-1 host gene (non-protein coding) |
| chr2_-_228497888 | 0.02 |
ENST00000264387.4
ENST00000409066.1 |
C2orf83
|
chromosome 2 open reading frame 83 |
| chr19_+_58694396 | 0.02 |
ENST00000326804.4
ENST00000345813.3 ENST00000424679.2 |
ZNF274
|
zinc finger protein 274 |
| chr1_-_207226313 | 0.02 |
ENST00000367084.1
|
YOD1
|
YOD1 deubiquitinase |
| chr17_+_67498396 | 0.02 |
ENST00000588110.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr9_-_107754034 | 0.02 |
ENST00000457720.1
|
RP11-217B7.3
|
RP11-217B7.3 |
| chr17_+_79651007 | 0.02 |
ENST00000572392.1
ENST00000577012.1 |
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate |
| chr12_-_10282836 | 0.02 |
ENST00000304084.8
ENST00000353231.5 ENST00000525605.1 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr7_+_100860949 | 0.02 |
ENST00000305105.2
|
ZNHIT1
|
zinc finger, HIT-type containing 1 |
| chr6_+_106988986 | 0.02 |
ENST00000457437.1
ENST00000535438.1 |
AIM1
|
absent in melanoma 1 |
| chr11_-_107729287 | 0.02 |
ENST00000375682.4
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr3_+_15643245 | 0.02 |
ENST00000303498.5
ENST00000437172.1 |
BTD
|
biotinidase |
| chr17_+_62223320 | 0.02 |
ENST00000580828.1
ENST00000582965.1 |
SNORA76
|
small nucleolar RNA, H/ACA box 76 |
| chr17_+_74846230 | 0.02 |
ENST00000592919.1
|
LINC00868
|
long intergenic non-protein coding RNA 868 |
| chr19_+_9296279 | 0.02 |
ENST00000344248.2
|
OR7D2
|
olfactory receptor, family 7, subfamily D, member 2 |
| chr19_-_10491234 | 0.02 |
ENST00000524462.1
ENST00000531836.1 ENST00000525621.1 |
TYK2
|
tyrosine kinase 2 |
| chr12_-_86650077 | 0.02 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr8_-_65730127 | 0.02 |
ENST00000522106.1
|
RP11-1D12.2
|
RP11-1D12.2 |
| chr19_-_14064114 | 0.02 |
ENST00000585607.1
ENST00000538517.2 ENST00000587458.1 ENST00000538371.2 |
PODNL1
|
podocan-like 1 |
| chr15_-_78913628 | 0.02 |
ENST00000348639.3
|
CHRNA3
|
cholinergic receptor, nicotinic, alpha 3 (neuronal) |
| chr4_-_118006697 | 0.02 |
ENST00000310754.4
|
TRAM1L1
|
translocation associated membrane protein 1-like 1 |
| chr4_-_48116540 | 0.02 |
ENST00000506073.1
|
TXK
|
TXK tyrosine kinase |
| chr10_+_90562705 | 0.02 |
ENST00000539337.1
|
LIPM
|
lipase, family member M |
| chr4_-_113558014 | 0.02 |
ENST00000503172.1
ENST00000505019.1 ENST00000309071.5 |
C4orf21
|
chromosome 4 open reading frame 21 |
| chr19_+_2785458 | 0.02 |
ENST00000307741.6
ENST00000585338.1 |
THOP1
|
thimet oligopeptidase 1 |
| chr11_+_65265141 | 0.02 |
ENST00000534336.1
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr12_+_7014064 | 0.02 |
ENST00000443597.2
|
LRRC23
|
leucine rich repeat containing 23 |
| chr7_+_100551239 | 0.02 |
ENST00000319509.7
|
MUC3A
|
mucin 3A, cell surface associated |
| chr3_-_9885626 | 0.02 |
ENST00000424438.1
ENST00000433555.1 ENST00000427174.1 ENST00000418713.1 ENST00000433535.2 ENST00000383820.5 ENST00000433972.1 |
RPUSD3
|
RNA pseudouridylate synthase domain containing 3 |
| chr2_+_161993465 | 0.02 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr1_-_1709845 | 0.01 |
ENST00000341426.5
ENST00000344463.4 |
NADK
|
NAD kinase |
| chrX_+_108779004 | 0.01 |
ENST00000218004.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SHOX2_HOXC5
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.0 | 0.2 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.1 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.0 | 0.0 | GO:0017143 | insecticide metabolic process(GO:0017143) cellular response to luteinizing hormone stimulus(GO:0071373) |
| 0.0 | 0.0 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.0 | 0.0 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |