Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for SHOX
Z-value: 0.60
Transcription factors associated with SHOX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SHOX
|
ENSG00000185960.8 | short stature homeobox |
|
SHOX
|
ENSGR0000185960.8 | short stature homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SHOX | hg19_v2_chrX_+_591524_591542 | 0.85 | 1.5e-01 | Click! |
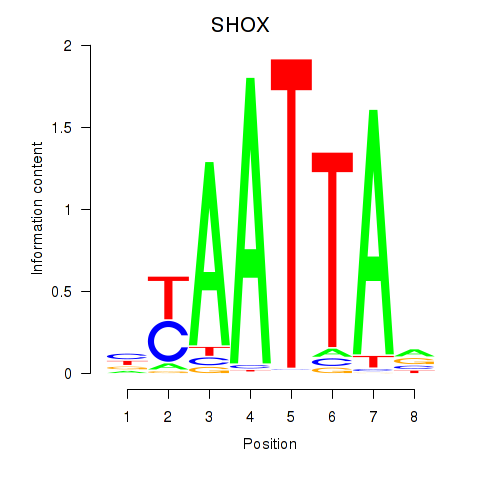
Activity profile of SHOX motif
Sorted Z-values of SHOX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_167191814 | 0.56 |
ENST00000466903.1
ENST00000264677.4 |
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2 |
| chr9_-_3469181 | 0.40 |
ENST00000366116.2
|
AL365202.1
|
Uncharacterized protein |
| chr7_-_99716914 | 0.39 |
ENST00000431404.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr3_-_178103144 | 0.39 |
ENST00000417383.1
ENST00000418585.1 ENST00000411727.1 ENST00000439810.1 |
RP11-33A14.1
|
RP11-33A14.1 |
| chr5_+_136070614 | 0.36 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1 |
| chr12_+_52695617 | 0.35 |
ENST00000293525.5
|
KRT86
|
keratin 86 |
| chr17_-_46690839 | 0.31 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr5_-_56778635 | 0.30 |
ENST00000423391.1
|
ACTBL2
|
actin, beta-like 2 |
| chr6_+_155538093 | 0.30 |
ENST00000462408.2
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr2_+_162272605 | 0.27 |
ENST00000389554.3
|
TBR1
|
T-box, brain, 1 |
| chr2_-_99871570 | 0.26 |
ENST00000333017.2
ENST00000409679.1 ENST00000423306.1 |
LYG2
|
lysozyme G-like 2 |
| chr3_-_27764190 | 0.24 |
ENST00000537516.1
|
EOMES
|
eomesodermin |
| chr11_+_35222629 | 0.23 |
ENST00000526553.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr17_-_19015945 | 0.23 |
ENST00000573866.2
|
SNORD3D
|
small nucleolar RNA, C/D box 3D |
| chr12_-_89746173 | 0.22 |
ENST00000308385.6
|
DUSP6
|
dual specificity phosphatase 6 |
| chr12_-_89746264 | 0.22 |
ENST00000548755.1
|
DUSP6
|
dual specificity phosphatase 6 |
| chr5_+_101569696 | 0.22 |
ENST00000597120.1
|
AC008948.1
|
AC008948.1 |
| chr6_-_39693111 | 0.20 |
ENST00000373215.3
ENST00000538893.1 ENST00000287152.7 ENST00000373216.3 |
KIF6
|
kinesin family member 6 |
| chr14_-_36988882 | 0.19 |
ENST00000498187.2
|
NKX2-1
|
NK2 homeobox 1 |
| chr10_+_35484793 | 0.19 |
ENST00000488741.1
ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chr12_+_78359999 | 0.18 |
ENST00000550503.1
|
NAV3
|
neuron navigator 3 |
| chr7_-_14026063 | 0.18 |
ENST00000443608.1
ENST00000438956.1 |
ETV1
|
ets variant 1 |
| chr19_+_48949087 | 0.18 |
ENST00000598711.1
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chr17_+_56833184 | 0.18 |
ENST00000308249.2
|
PPM1E
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E |
| chr19_-_51523412 | 0.17 |
ENST00000391805.1
ENST00000599077.1 |
KLK10
|
kallikrein-related peptidase 10 |
| chr12_-_10282742 | 0.17 |
ENST00000298523.5
ENST00000396484.2 ENST00000310002.4 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr6_+_4087664 | 0.16 |
ENST00000430835.2
|
C6orf201
|
chromosome 6 open reading frame 201 |
| chr18_+_57567180 | 0.16 |
ENST00000316660.6
ENST00000269518.9 |
PMAIP1
|
phorbol-12-myristate-13-acetate-induced protein 1 |
| chr12_-_102591604 | 0.16 |
ENST00000329406.4
|
PMCH
|
pro-melanin-concentrating hormone |
| chr13_+_78315295 | 0.16 |
ENST00000351546.3
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr9_+_82188077 | 0.16 |
ENST00000425506.1
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr2_+_45168875 | 0.15 |
ENST00000260653.3
|
SIX3
|
SIX homeobox 3 |
| chr1_+_12042015 | 0.15 |
ENST00000412236.1
|
MFN2
|
mitofusin 2 |
| chr8_+_107738343 | 0.15 |
ENST00000521592.1
|
OXR1
|
oxidation resistance 1 |
| chr21_+_17909594 | 0.15 |
ENST00000441820.1
ENST00000602280.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr15_+_63188009 | 0.14 |
ENST00000557900.1
|
RP11-1069G10.2
|
RP11-1069G10.2 |
| chr3_-_98235962 | 0.14 |
ENST00000513873.1
|
CLDND1
|
claudin domain containing 1 |
| chr2_+_179318295 | 0.14 |
ENST00000442710.1
|
DFNB59
|
deafness, autosomal recessive 59 |
| chr3_-_108248169 | 0.14 |
ENST00000273353.3
|
MYH15
|
myosin, heavy chain 15 |
| chr3_-_71777824 | 0.14 |
ENST00000469524.1
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr7_-_14026123 | 0.14 |
ENST00000420159.2
ENST00000399357.3 ENST00000403527.1 |
ETV1
|
ets variant 1 |
| chr5_+_174151536 | 0.13 |
ENST00000239243.6
ENST00000507785.1 |
MSX2
|
msh homeobox 2 |
| chr7_+_116660246 | 0.13 |
ENST00000434836.1
ENST00000393443.1 ENST00000465133.1 ENST00000477742.1 ENST00000393447.4 ENST00000393444.3 |
ST7
|
suppression of tumorigenicity 7 |
| chr1_+_153747746 | 0.13 |
ENST00000368661.3
|
SLC27A3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr3_+_141106643 | 0.13 |
ENST00000514251.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr2_-_190927447 | 0.13 |
ENST00000260950.4
|
MSTN
|
myostatin |
| chr7_+_115862858 | 0.13 |
ENST00000393481.2
|
TES
|
testis derived transcript (3 LIM domains) |
| chr21_-_35284635 | 0.13 |
ENST00000429238.1
|
AP000304.12
|
AP000304.12 |
| chr14_-_98444461 | 0.13 |
ENST00000499006.2
|
C14orf64
|
chromosome 14 open reading frame 64 |
| chr10_+_94451574 | 0.13 |
ENST00000492654.2
|
HHEX
|
hematopoietically expressed homeobox |
| chr11_-_31531121 | 0.12 |
ENST00000532287.1
ENST00000526776.1 ENST00000534812.1 ENST00000529749.1 ENST00000278200.1 ENST00000530023.1 ENST00000533642.1 |
IMMP1L
|
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr11_-_117747434 | 0.12 |
ENST00000529335.2
ENST00000530956.1 ENST00000260282.4 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr19_-_53758094 | 0.12 |
ENST00000601828.1
ENST00000598513.1 ENST00000599012.1 ENST00000333952.4 ENST00000598806.1 |
ZNF677
|
zinc finger protein 677 |
| chr6_+_52535878 | 0.12 |
ENST00000211314.4
|
TMEM14A
|
transmembrane protein 14A |
| chr18_-_500692 | 0.12 |
ENST00000400256.3
|
COLEC12
|
collectin sub-family member 12 |
| chr2_+_66918558 | 0.12 |
ENST00000435389.1
ENST00000428590.1 ENST00000412944.1 |
AC007392.3
|
AC007392.3 |
| chrX_+_108779004 | 0.12 |
ENST00000218004.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr12_+_49621658 | 0.12 |
ENST00000541364.1
|
TUBA1C
|
tubulin, alpha 1c |
| chr8_+_107738240 | 0.12 |
ENST00000449762.2
ENST00000297447.6 |
OXR1
|
oxidation resistance 1 |
| chr17_+_47448102 | 0.12 |
ENST00000576461.1
|
RP11-81K2.1
|
Uncharacterized protein |
| chr4_-_170897045 | 0.11 |
ENST00000508313.1
|
RP11-205M3.3
|
RP11-205M3.3 |
| chr3_-_180397256 | 0.11 |
ENST00000442201.2
|
CCDC39
|
coiled-coil domain containing 39 |
| chr9_-_69229650 | 0.11 |
ENST00000416428.1
|
CBWD6
|
COBW domain containing 6 |
| chr20_+_18447771 | 0.11 |
ENST00000377603.4
|
POLR3F
|
polymerase (RNA) III (DNA directed) polypeptide F, 39 kDa |
| chr20_+_2276639 | 0.11 |
ENST00000381458.5
|
TGM3
|
transglutaminase 3 |
| chr5_+_133562095 | 0.11 |
ENST00000602919.1
|
CTD-2410N18.3
|
CTD-2410N18.3 |
| chr14_-_54425475 | 0.11 |
ENST00000559642.1
|
BMP4
|
bone morphogenetic protein 4 |
| chr12_-_57328187 | 0.10 |
ENST00000293502.1
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C, member 7 |
| chr1_-_45956822 | 0.10 |
ENST00000372086.3
ENST00000341771.6 |
TESK2
|
testis-specific kinase 2 |
| chrX_+_22050546 | 0.10 |
ENST00000379374.4
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked |
| chr3_+_129247479 | 0.10 |
ENST00000296271.3
|
RHO
|
rhodopsin |
| chr19_-_14064114 | 0.10 |
ENST00000585607.1
ENST00000538517.2 ENST00000587458.1 ENST00000538371.2 |
PODNL1
|
podocan-like 1 |
| chr20_+_18488137 | 0.10 |
ENST00000450074.1
ENST00000262544.2 ENST00000336714.3 ENST00000377475.3 |
SEC23B
|
Sec23 homolog B (S. cerevisiae) |
| chr19_+_13049413 | 0.10 |
ENST00000316448.5
ENST00000588454.1 |
CALR
|
calreticulin |
| chr14_+_101359265 | 0.10 |
ENST00000599197.1
|
AL117190.3
|
Esophagus cancer-related gene-2 interaction susceptibility protein; Uncharacterized protein |
| chr1_-_151762943 | 0.10 |
ENST00000368825.3
ENST00000368823.1 ENST00000458431.2 ENST00000368827.6 ENST00000368824.3 |
TDRKH
|
tudor and KH domain containing |
| chr4_+_90033968 | 0.10 |
ENST00000317005.2
|
TIGD2
|
tigger transposable element derived 2 |
| chr2_-_136678123 | 0.10 |
ENST00000422708.1
|
DARS
|
aspartyl-tRNA synthetase |
| chr1_+_186265399 | 0.10 |
ENST00000367486.3
ENST00000367484.3 ENST00000533951.1 ENST00000367482.4 ENST00000367483.4 ENST00000367485.4 ENST00000445192.2 |
PRG4
|
proteoglycan 4 |
| chr6_+_5261225 | 0.10 |
ENST00000324331.6
|
FARS2
|
phenylalanyl-tRNA synthetase 2, mitochondrial |
| chr15_+_101420028 | 0.09 |
ENST00000557963.1
ENST00000346623.6 |
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3 |
| chr4_-_105416039 | 0.09 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr1_+_214161854 | 0.09 |
ENST00000435016.1
|
PROX1
|
prospero homeobox 1 |
| chr8_+_107460147 | 0.09 |
ENST00000442977.2
|
OXR1
|
oxidation resistance 1 |
| chrX_-_19817869 | 0.09 |
ENST00000379698.4
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr2_-_145277569 | 0.09 |
ENST00000303660.4
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr7_-_44580861 | 0.09 |
ENST00000546276.1
ENST00000289547.4 ENST00000381160.3 ENST00000423141.1 |
NPC1L1
|
NPC1-like 1 |
| chr7_-_137028534 | 0.09 |
ENST00000348225.2
|
PTN
|
pleiotrophin |
| chr1_+_153746683 | 0.09 |
ENST00000271857.2
|
SLC27A3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr16_-_28192360 | 0.09 |
ENST00000570033.1
|
XPO6
|
exportin 6 |
| chr12_-_52761262 | 0.09 |
ENST00000257901.3
|
KRT85
|
keratin 85 |
| chr7_-_137028498 | 0.08 |
ENST00000393083.2
|
PTN
|
pleiotrophin |
| chr11_+_112041253 | 0.08 |
ENST00000532612.1
|
AP002884.3
|
AP002884.3 |
| chr1_-_242612779 | 0.08 |
ENST00000427495.1
|
PLD5
|
phospholipase D family, member 5 |
| chr17_+_18086392 | 0.08 |
ENST00000541285.1
|
ALKBH5
|
alkB, alkylation repair homolog 5 (E. coli) |
| chr11_+_128563652 | 0.08 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr17_+_39405939 | 0.08 |
ENST00000334109.2
|
KRTAP9-4
|
keratin associated protein 9-4 |
| chrX_+_591524 | 0.08 |
ENST00000554971.1
ENST00000381575.1 |
SHOX
|
short stature homeobox |
| chr20_+_57414743 | 0.08 |
ENST00000313949.7
|
GNAS
|
GNAS complex locus |
| chr1_-_45956868 | 0.08 |
ENST00000451835.2
|
TESK2
|
testis-specific kinase 2 |
| chr6_-_116833500 | 0.08 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr3_+_151591422 | 0.08 |
ENST00000362032.5
|
SUCNR1
|
succinate receptor 1 |
| chr10_+_115511213 | 0.08 |
ENST00000361048.1
|
PLEKHS1
|
pleckstrin homology domain containing, family S member 1 |
| chr4_+_40201954 | 0.08 |
ENST00000511121.1
|
RHOH
|
ras homolog family member H |
| chr18_-_33709268 | 0.08 |
ENST00000269187.5
ENST00000590986.1 ENST00000440549.2 |
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr6_-_31107127 | 0.08 |
ENST00000259845.4
|
PSORS1C2
|
psoriasis susceptibility 1 candidate 2 |
| chr17_+_41363854 | 0.08 |
ENST00000588693.1
ENST00000588659.1 ENST00000541594.1 ENST00000536052.1 ENST00000331615.3 |
TMEM106A
|
transmembrane protein 106A |
| chr5_-_96478466 | 0.08 |
ENST00000274382.4
|
LIX1
|
Lix1 homolog (chicken) |
| chr21_+_17443434 | 0.08 |
ENST00000400178.2
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr12_-_56694142 | 0.08 |
ENST00000550655.1
ENST00000548567.1 ENST00000551430.2 ENST00000351328.3 |
CS
|
citrate synthase |
| chr12_-_31478428 | 0.08 |
ENST00000543615.1
|
FAM60A
|
family with sequence similarity 60, member A |
| chr17_-_48785216 | 0.08 |
ENST00000285243.6
|
ANKRD40
|
ankyrin repeat domain 40 |
| chr10_+_90660832 | 0.07 |
ENST00000371924.1
|
STAMBPL1
|
STAM binding protein-like 1 |
| chr19_-_51522955 | 0.07 |
ENST00000358789.3
|
KLK10
|
kallikrein-related peptidase 10 |
| chrX_+_107288239 | 0.07 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr10_-_71169031 | 0.07 |
ENST00000373307.1
|
TACR2
|
tachykinin receptor 2 |
| chr7_+_138943265 | 0.07 |
ENST00000483726.1
|
UBN2
|
ubinuclein 2 |
| chr6_+_130339710 | 0.07 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr3_+_141106458 | 0.07 |
ENST00000509883.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr15_+_36887069 | 0.07 |
ENST00000566807.1
ENST00000567389.1 ENST00000562877.1 |
C15orf41
|
chromosome 15 open reading frame 41 |
| chr9_+_22646189 | 0.07 |
ENST00000436786.1
|
RP11-399D6.2
|
RP11-399D6.2 |
| chr1_+_198608146 | 0.07 |
ENST00000367376.2
ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr14_+_67831576 | 0.07 |
ENST00000555876.1
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa |
| chr2_-_145278475 | 0.07 |
ENST00000558170.2
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr4_+_146402346 | 0.07 |
ENST00000514778.1
ENST00000507594.1 |
SMAD1
|
SMAD family member 1 |
| chr15_+_80351977 | 0.07 |
ENST00000559157.1
ENST00000561012.1 ENST00000564367.1 ENST00000558494.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr7_+_135611542 | 0.07 |
ENST00000416501.1
|
AC015987.2
|
AC015987.2 |
| chr9_-_16728161 | 0.07 |
ENST00000603713.1
ENST00000603313.1 |
BNC2
|
basonuclin 2 |
| chr12_+_20963647 | 0.07 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr13_+_78315528 | 0.07 |
ENST00000496045.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr3_+_69928256 | 0.07 |
ENST00000394355.2
|
MITF
|
microphthalmia-associated transcription factor |
| chr12_-_16759440 | 0.06 |
ENST00000537304.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chrX_+_107288197 | 0.06 |
ENST00000415430.3
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr1_+_180601139 | 0.06 |
ENST00000367590.4
ENST00000367589.3 |
XPR1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr7_+_138145076 | 0.06 |
ENST00000343526.4
|
TRIM24
|
tripartite motif containing 24 |
| chr8_+_117950422 | 0.06 |
ENST00000378279.3
|
AARD
|
alanine and arginine rich domain containing protein |
| chr1_+_214161272 | 0.06 |
ENST00000498508.2
ENST00000366958.4 |
PROX1
|
prospero homeobox 1 |
| chr15_-_55562479 | 0.06 |
ENST00000564609.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr1_+_160370344 | 0.06 |
ENST00000368061.2
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr7_+_116502527 | 0.06 |
ENST00000361183.3
|
CAPZA2
|
capping protein (actin filament) muscle Z-line, alpha 2 |
| chr19_+_30719410 | 0.06 |
ENST00000585628.1
ENST00000591488.1 |
ZNF536
|
zinc finger protein 536 |
| chr15_+_80351910 | 0.06 |
ENST00000261749.6
ENST00000561060.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr4_+_72204755 | 0.06 |
ENST00000512686.1
ENST00000340595.3 |
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr1_-_224624730 | 0.06 |
ENST00000445239.1
|
WDR26
|
WD repeat domain 26 |
| chr20_-_45980621 | 0.06 |
ENST00000446894.1
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr13_+_36050881 | 0.06 |
ENST00000537702.1
|
NBEA
|
neurobeachin |
| chr1_-_232651312 | 0.06 |
ENST00000262861.4
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2 |
| chr1_-_151762900 | 0.06 |
ENST00000440583.2
|
TDRKH
|
tudor and KH domain containing |
| chr12_-_110937351 | 0.06 |
ENST00000552130.2
|
VPS29
|
vacuolar protein sorting 29 homolog (S. cerevisiae) |
| chr20_+_55967129 | 0.06 |
ENST00000371219.2
|
RBM38
|
RNA binding motif protein 38 |
| chr11_-_117748138 | 0.06 |
ENST00000527717.1
|
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr6_-_127780510 | 0.06 |
ENST00000487331.2
ENST00000483725.3 |
KIAA0408
|
KIAA0408 |
| chr21_+_17443521 | 0.06 |
ENST00000456342.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr6_-_138539627 | 0.06 |
ENST00000527246.2
|
PBOV1
|
prostate and breast cancer overexpressed 1 |
| chr14_+_32798547 | 0.06 |
ENST00000557354.1
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr16_+_103816 | 0.06 |
ENST00000383018.3
ENST00000417493.1 |
SNRNP25
|
small nuclear ribonucleoprotein 25kDa (U11/U12) |
| chr6_+_26365443 | 0.06 |
ENST00000527422.1
ENST00000356386.2 ENST00000396934.3 ENST00000377708.2 ENST00000396948.1 ENST00000508906.2 |
BTN3A2
|
butyrophilin, subfamily 3, member A2 |
| chr11_+_123430948 | 0.06 |
ENST00000529432.1
ENST00000534764.1 |
GRAMD1B
|
GRAM domain containing 1B |
| chr1_-_101360331 | 0.05 |
ENST00000416479.1
ENST00000370113.3 |
EXTL2
|
exostosin-like glycosyltransferase 2 |
| chr3_+_156393349 | 0.05 |
ENST00000473702.1
|
TIPARP
|
TCDD-inducible poly(ADP-ribose) polymerase |
| chr15_-_55562582 | 0.05 |
ENST00000396307.2
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr1_+_109265033 | 0.05 |
ENST00000445274.1
|
FNDC7
|
fibronectin type III domain containing 7 |
| chr14_-_54423529 | 0.05 |
ENST00000245451.4
ENST00000559087.1 |
BMP4
|
bone morphogenetic protein 4 |
| chr18_-_44181442 | 0.05 |
ENST00000398722.4
|
LOXHD1
|
lipoxygenase homology domains 1 |
| chr2_-_200335983 | 0.05 |
ENST00000457245.1
|
SATB2
|
SATB homeobox 2 |
| chr12_-_122018346 | 0.05 |
ENST00000377069.4
|
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr11_-_111794446 | 0.05 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr11_-_117747607 | 0.05 |
ENST00000540359.1
ENST00000539526.1 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr3_-_178984759 | 0.05 |
ENST00000349697.2
ENST00000497599.1 |
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr12_-_10282836 | 0.05 |
ENST00000304084.8
ENST00000353231.5 ENST00000525605.1 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr4_+_95916947 | 0.05 |
ENST00000506363.1
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr1_+_206557366 | 0.05 |
ENST00000414007.1
ENST00000419187.2 |
SRGAP2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr12_-_112123524 | 0.05 |
ENST00000327551.6
|
BRAP
|
BRCA1 associated protein |
| chr15_+_48483736 | 0.05 |
ENST00000417307.2
ENST00000559641.1 |
CTXN2
SLC12A1
|
cortexin 2 solute carrier family 12 (sodium/potassium/chloride transporter), member 1 |
| chrX_-_110655306 | 0.05 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr21_+_17442799 | 0.05 |
ENST00000602580.1
ENST00000458468.1 ENST00000602935.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr2_-_151395525 | 0.05 |
ENST00000439275.1
|
RND3
|
Rho family GTPase 3 |
| chr7_+_77428149 | 0.05 |
ENST00000415251.2
ENST00000275575.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr17_-_46716647 | 0.05 |
ENST00000608940.1
|
RP11-357H14.17
|
RP11-357H14.17 |
| chr11_-_117747327 | 0.05 |
ENST00000584230.1
ENST00000527429.1 ENST00000584394.1 ENST00000532984.1 |
FXYD6
FXYD6-FXYD2
|
FXYD domain containing ion transport regulator 6 FXYD6-FXYD2 readthrough |
| chr7_+_141408153 | 0.05 |
ENST00000397541.2
|
WEE2
|
WEE1 homolog 2 (S. pombe) |
| chr21_-_35899113 | 0.05 |
ENST00000492600.1
ENST00000481448.1 ENST00000381132.2 |
RCAN1
|
regulator of calcineurin 1 |
| chr17_+_59489112 | 0.05 |
ENST00000335108.2
|
C17orf82
|
chromosome 17 open reading frame 82 |
| chr14_-_64194745 | 0.05 |
ENST00000247225.6
|
SGPP1
|
sphingosine-1-phosphate phosphatase 1 |
| chr9_-_123639445 | 0.05 |
ENST00000312189.6
|
PHF19
|
PHD finger protein 19 |
| chr7_-_14028488 | 0.05 |
ENST00000405358.4
|
ETV1
|
ets variant 1 |
| chr8_-_17533838 | 0.05 |
ENST00000400046.1
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr12_-_28122980 | 0.05 |
ENST00000395868.3
ENST00000534890.1 |
PTHLH
|
parathyroid hormone-like hormone |
| chr19_+_54641444 | 0.05 |
ENST00000221232.5
ENST00000358389.3 |
CNOT3
|
CCR4-NOT transcription complex, subunit 3 |
| chr6_+_55192267 | 0.05 |
ENST00000340465.2
|
GFRAL
|
GDNF family receptor alpha like |
| chr11_+_24518723 | 0.05 |
ENST00000336930.6
ENST00000529015.1 ENST00000533227.1 |
LUZP2
|
leucine zipper protein 2 |
| chr4_-_36245561 | 0.05 |
ENST00000506189.1
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr12_-_88974236 | 0.05 |
ENST00000228280.5
ENST00000552044.1 ENST00000357116.4 |
KITLG
|
KIT ligand |
| chr14_+_38065052 | 0.04 |
ENST00000556845.1
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chr12_-_58135903 | 0.04 |
ENST00000257897.3
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr12_+_7014064 | 0.04 |
ENST00000443597.2
|
LRRC23
|
leucine rich repeat containing 23 |
| chr2_-_74618907 | 0.04 |
ENST00000421392.1
ENST00000437375.1 |
DCTN1
|
dynactin 1 |
| chr11_+_75526212 | 0.04 |
ENST00000356136.3
|
UVRAG
|
UV radiation resistance associated |
| chr14_+_74815116 | 0.04 |
ENST00000256362.4
|
VRTN
|
vertebrae development associated |
| chr6_+_153552455 | 0.04 |
ENST00000392385.2
|
AL590867.1
|
Uncharacterized protein; cDNA FLJ59044, highly similar to LINE-1 reverse transcriptase homolog |
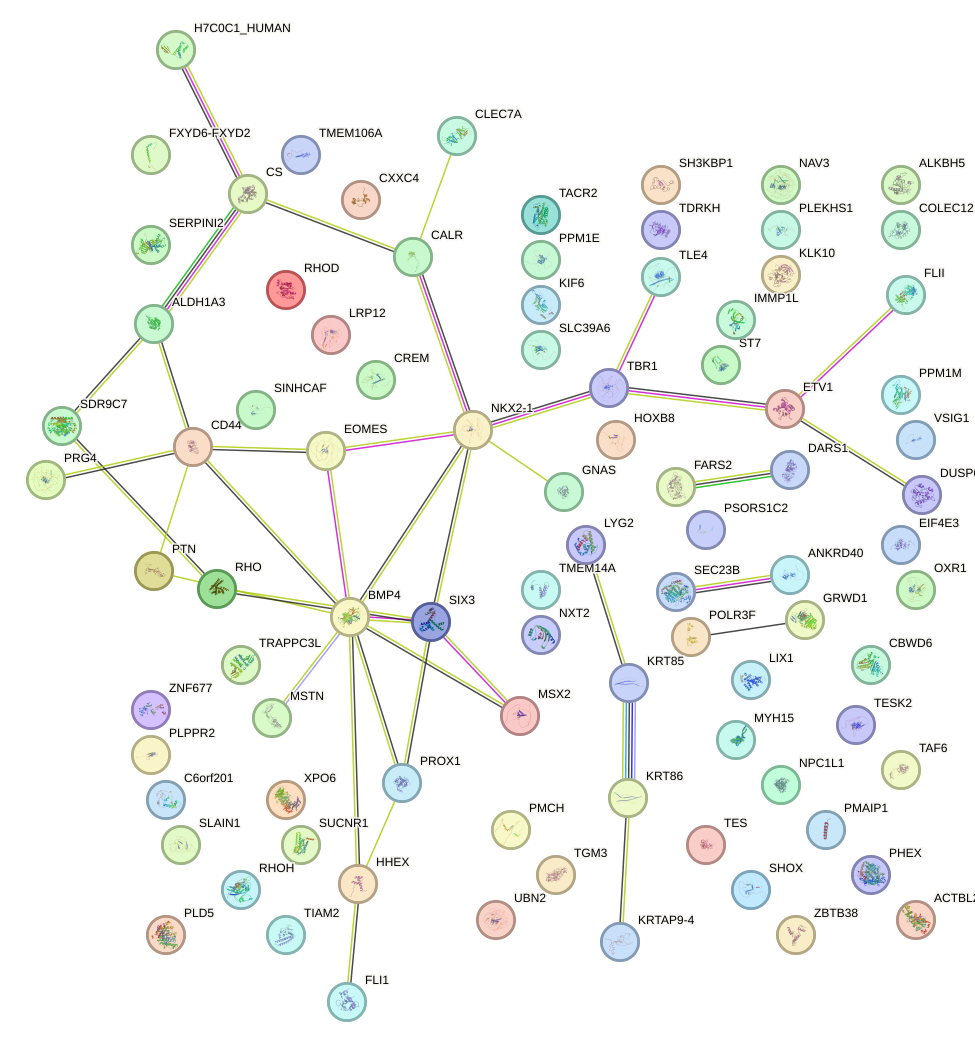
Network of associatons between targets according to the STRING database.
First level regulatory network of SHOX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.1 | 0.1 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.1 | 0.2 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 0.1 | 0.2 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.0 | 0.1 | GO:0014016 | neuroblast differentiation(GO:0014016) forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.1 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.0 | 0.1 | GO:0051795 | positive regulation of catagen(GO:0051795) activation of meiosis(GO:0090427) |
| 0.0 | 0.3 | GO:0021764 | amygdala development(GO:0021764) |
| 0.0 | 0.1 | GO:1902725 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.3 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.0 | 0.3 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.2 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.2 | GO:0061151 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.0 | 0.2 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0060488 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.0 | 0.1 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.1 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.1 | GO:0033685 | negative regulation of luteinizing hormone secretion(GO:0033685) |
| 0.0 | 0.1 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.4 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.2 | GO:1903056 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.0 | 0.1 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.0 | 0.1 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.1 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.3 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.4 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.0 | GO:0033023 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.0 | 0.0 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.3 | GO:0007625 | grooming behavior(GO:0007625) |
| 0.0 | 0.1 | GO:0002501 | MHC protein complex assembly(GO:0002396) peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.0 | 0.1 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.1 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.1 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.0 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.2 | GO:1902237 | positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902237) |
| 0.0 | 0.0 | GO:0003292 | cardiac septum cell differentiation(GO:0003292) atrioventricular node cell differentiation(GO:0060922) atrioventricular node cell development(GO:0060928) |
| 0.0 | 0.1 | GO:0061734 | parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.2 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.1 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.3 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.2 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.3 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.0 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.3 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.0 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |