Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
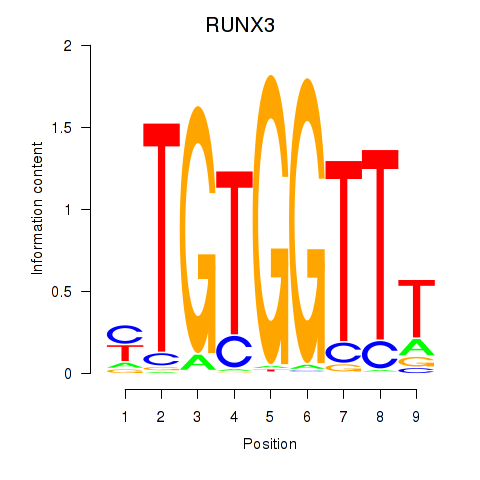
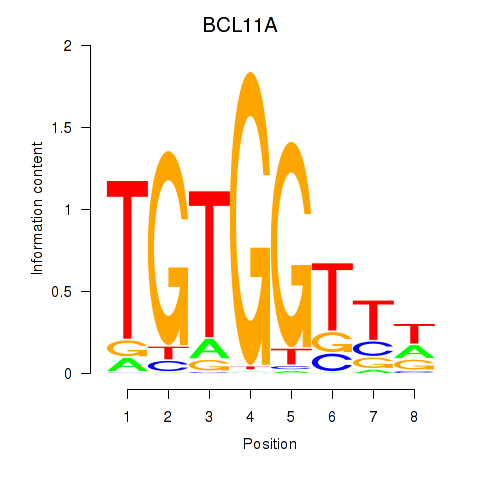
Results for RUNX3_BCL11A
Z-value: 0.36
Transcription factors associated with RUNX3_BCL11A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RUNX3
|
ENSG00000020633.14 | RUNX family transcription factor 3 |
|
BCL11A
|
ENSG00000119866.16 | BAF chromatin remodeling complex subunit BCL11A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RUNX3 | hg19_v2_chr1_-_25256368_25256476 | -0.93 | 7.5e-02 | Click! |
| BCL11A | hg19_v2_chr2_-_60780702_60780742 | 0.52 | 4.8e-01 | Click! |
Activity profile of RUNX3_BCL11A motif
Sorted Z-values of RUNX3_BCL11A motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_156308245 | 0.26 |
ENST00000368253.2
ENST00000470342.1 ENST00000368254.1 |
TSACC
|
TSSK6 activating co-chaperone |
| chr1_-_235098935 | 0.24 |
ENST00000423175.1
|
RP11-443B7.1
|
RP11-443B7.1 |
| chr2_+_136499287 | 0.23 |
ENST00000415164.1
|
UBXN4
|
UBX domain protein 4 |
| chr1_+_145727681 | 0.18 |
ENST00000417171.1
ENST00000451928.2 |
PDZK1
|
PDZ domain containing 1 |
| chr1_-_168464875 | 0.16 |
ENST00000422253.1
|
RP5-968D22.3
|
RP5-968D22.3 |
| chr1_+_156308403 | 0.16 |
ENST00000481479.1
ENST00000368252.1 ENST00000466306.1 ENST00000368251.1 |
TSACC
|
TSSK6 activating co-chaperone |
| chr6_+_45296391 | 0.15 |
ENST00000371436.6
ENST00000576263.1 |
RUNX2
|
runt-related transcription factor 2 |
| chr11_-_1606513 | 0.12 |
ENST00000382171.2
|
KRTAP5-1
|
keratin associated protein 5-1 |
| chr17_+_76037081 | 0.11 |
ENST00000588549.1
|
TNRC6C
|
trinucleotide repeat containing 6C |
| chr6_+_27925019 | 0.10 |
ENST00000244623.1
|
OR2B6
|
olfactory receptor, family 2, subfamily B, member 6 |
| chr1_+_156211753 | 0.10 |
ENST00000368272.4
|
BGLAP
|
bone gamma-carboxyglutamate (gla) protein |
| chr5_-_39270725 | 0.09 |
ENST00000512138.1
ENST00000512982.1 ENST00000540520.1 |
FYB
|
FYN binding protein |
| chr3_+_49977894 | 0.09 |
ENST00000433811.1
|
RBM6
|
RNA binding motif protein 6 |
| chr14_-_23791484 | 0.09 |
ENST00000594872.1
|
AL049829.1
|
Uncharacterized protein |
| chr3_+_142342240 | 0.09 |
ENST00000497199.1
|
PLS1
|
plastin 1 |
| chr19_-_50868882 | 0.08 |
ENST00000598915.1
|
NAPSA
|
napsin A aspartic peptidase |
| chrX_-_49121165 | 0.08 |
ENST00000376207.4
ENST00000376199.2 |
FOXP3
|
forkhead box P3 |
| chrX_+_155227371 | 0.08 |
ENST00000369423.2
ENST00000540897.1 |
IL9R
|
interleukin 9 receptor |
| chr12_-_96793142 | 0.08 |
ENST00000552262.1
ENST00000551816.1 ENST00000552496.1 |
CDK17
|
cyclin-dependent kinase 17 |
| chrX_+_155227246 | 0.08 |
ENST00000244174.5
ENST00000424344.3 |
IL9R
|
interleukin 9 receptor |
| chr1_+_110453109 | 0.07 |
ENST00000525659.1
|
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr15_+_86098670 | 0.07 |
ENST00000558811.1
|
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr2_+_44001172 | 0.07 |
ENST00000260605.8
ENST00000406852.3 ENST00000443170.3 ENST00000398823.2 ENST00000605786.1 |
DYNC2LI1
|
dynein, cytoplasmic 2, light intermediate chain 1 |
| chr3_+_136676707 | 0.07 |
ENST00000329582.4
|
IL20RB
|
interleukin 20 receptor beta |
| chr17_-_46692457 | 0.07 |
ENST00000468443.1
|
HOXB8
|
homeobox B8 |
| chr3_+_49977440 | 0.07 |
ENST00000442092.1
ENST00000266022.4 ENST00000443081.1 |
RBM6
|
RNA binding motif protein 6 |
| chr1_+_44889697 | 0.07 |
ENST00000443020.2
|
RNF220
|
ring finger protein 220 |
| chr7_+_70229899 | 0.07 |
ENST00000443672.1
|
AUTS2
|
autism susceptibility candidate 2 |
| chr17_+_9745786 | 0.07 |
ENST00000304773.5
|
GLP2R
|
glucagon-like peptide 2 receptor |
| chr11_+_60739249 | 0.07 |
ENST00000542157.1
ENST00000433107.2 ENST00000452451.2 ENST00000352009.5 |
CD6
|
CD6 molecule |
| chr15_+_58724184 | 0.07 |
ENST00000433326.2
|
LIPC
|
lipase, hepatic |
| chr19_-_8567505 | 0.07 |
ENST00000600262.1
|
PRAM1
|
PML-RARA regulated adaptor molecule 1 |
| chrX_+_48542168 | 0.06 |
ENST00000376701.4
|
WAS
|
Wiskott-Aldrich syndrome |
| chr17_-_73178599 | 0.06 |
ENST00000578238.1
|
SUMO2
|
small ubiquitin-like modifier 2 |
| chr14_-_35183755 | 0.06 |
ENST00000555765.1
|
CFL2
|
cofilin 2 (muscle) |
| chr6_-_52109335 | 0.06 |
ENST00000336123.4
|
IL17F
|
interleukin 17F |
| chr18_+_5238549 | 0.06 |
ENST00000580684.1
|
LINC00667
|
long intergenic non-protein coding RNA 667 |
| chr5_+_135496675 | 0.06 |
ENST00000507637.1
|
SMAD5
|
SMAD family member 5 |
| chr5_+_67535647 | 0.06 |
ENST00000520675.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr22_+_40440804 | 0.06 |
ENST00000441751.1
ENST00000301923.9 |
TNRC6B
|
trinucleotide repeat containing 6B |
| chr2_-_111291587 | 0.06 |
ENST00000437167.1
|
RGPD6
|
RANBP2-like and GRIP domain containing 6 |
| chrX_+_119005399 | 0.06 |
ENST00000371437.4
|
NDUFA1
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 1, 7.5kDa |
| chr17_-_73840614 | 0.06 |
ENST00000586108.1
|
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr2_+_74741569 | 0.06 |
ENST00000233638.7
|
TLX2
|
T-cell leukemia homeobox 2 |
| chr8_-_11325047 | 0.06 |
ENST00000531804.1
|
FAM167A
|
family with sequence similarity 167, member A |
| chr15_-_78358465 | 0.06 |
ENST00000435468.1
|
TBC1D2B
|
TBC1 domain family, member 2B |
| chr5_+_75699040 | 0.06 |
ENST00000274364.6
|
IQGAP2
|
IQ motif containing GTPase activating protein 2 |
| chr15_+_49715449 | 0.05 |
ENST00000560979.1
|
FGF7
|
fibroblast growth factor 7 |
| chr13_-_44735393 | 0.05 |
ENST00000400419.1
|
SMIM2
|
small integral membrane protein 2 |
| chr17_+_73629500 | 0.05 |
ENST00000375215.3
|
SMIM5
|
small integral membrane protein 5 |
| chr2_-_119605253 | 0.05 |
ENST00000295206.6
|
EN1
|
engrailed homeobox 1 |
| chr5_-_39203093 | 0.05 |
ENST00000515010.1
|
FYB
|
FYN binding protein |
| chr1_+_209929494 | 0.05 |
ENST00000367026.3
|
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr2_+_136499179 | 0.05 |
ENST00000272638.9
|
UBXN4
|
UBX domain protein 4 |
| chr9_+_125795788 | 0.05 |
ENST00000373643.5
|
RABGAP1
|
RAB GTPase activating protein 1 |
| chr17_-_42188598 | 0.05 |
ENST00000591714.1
|
HDAC5
|
histone deacetylase 5 |
| chr22_+_31488433 | 0.05 |
ENST00000455608.1
|
SMTN
|
smoothelin |
| chr11_+_62475130 | 0.05 |
ENST00000294117.5
|
GNG3
|
guanine nucleotide binding protein (G protein), gamma 3 |
| chr2_+_169658928 | 0.05 |
ENST00000317647.7
ENST00000445023.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr6_+_159071015 | 0.05 |
ENST00000360448.3
|
SYTL3
|
synaptotagmin-like 3 |
| chr5_+_55354822 | 0.05 |
ENST00000511861.1
|
CTD-2227I18.1
|
CTD-2227I18.1 |
| chr1_+_63788730 | 0.05 |
ENST00000371116.2
|
FOXD3
|
forkhead box D3 |
| chr17_-_74533963 | 0.05 |
ENST00000293230.5
|
CYGB
|
cytoglobin |
| chr5_+_75699149 | 0.05 |
ENST00000379730.3
|
IQGAP2
|
IQ motif containing GTPase activating protein 2 |
| chr3_-_33686925 | 0.05 |
ENST00000485378.2
ENST00000313350.6 ENST00000487200.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr17_-_39183452 | 0.05 |
ENST00000361883.5
|
KRTAP1-5
|
keratin associated protein 1-5 |
| chr1_+_196621002 | 0.05 |
ENST00000367429.4
ENST00000439155.2 |
CFH
|
complement factor H |
| chr4_-_156297949 | 0.05 |
ENST00000515654.1
|
MAP9
|
microtubule-associated protein 9 |
| chr5_-_94417314 | 0.05 |
ENST00000505208.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr1_+_196621156 | 0.05 |
ENST00000359637.2
|
CFH
|
complement factor H |
| chr2_-_61389168 | 0.05 |
ENST00000607743.1
ENST00000605902.1 |
RP11-493E12.1
|
RP11-493E12.1 |
| chr5_-_65018834 | 0.05 |
ENST00000506816.1
|
SGTB
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr5_-_138780159 | 0.04 |
ENST00000512473.1
ENST00000515581.1 ENST00000515277.1 |
DNAJC18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr2_-_175462934 | 0.04 |
ENST00000392546.2
ENST00000436221.1 |
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr19_-_42636543 | 0.04 |
ENST00000528894.4
ENST00000560804.2 ENST00000560558.1 ENST00000560398.1 ENST00000526816.2 |
POU2F2
|
POU class 2 homeobox 2 |
| chr2_+_97202480 | 0.04 |
ENST00000357485.3
|
ARID5A
|
AT rich interactive domain 5A (MRF1-like) |
| chr15_+_81225699 | 0.04 |
ENST00000560027.1
|
KIAA1199
|
KIAA1199 |
| chr5_-_94417339 | 0.04 |
ENST00000429576.2
ENST00000508509.1 ENST00000510732.1 |
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr7_+_18535321 | 0.04 |
ENST00000413380.1
ENST00000430454.1 |
HDAC9
|
histone deacetylase 9 |
| chr12_+_54694979 | 0.04 |
ENST00000552848.1
|
COPZ1
|
coatomer protein complex, subunit zeta 1 |
| chr17_-_36884451 | 0.04 |
ENST00000595377.1
|
AC006449.1
|
NS5ATP13TP1; Uncharacterized protein |
| chr1_+_23695680 | 0.04 |
ENST00000454117.1
ENST00000335648.3 ENST00000518821.1 ENST00000437367.2 |
C1orf213
|
chromosome 1 open reading frame 213 |
| chr5_-_138861926 | 0.04 |
ENST00000510817.1
|
TMEM173
|
transmembrane protein 173 |
| chr11_+_35198243 | 0.04 |
ENST00000528455.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr1_+_209929377 | 0.04 |
ENST00000400959.3
ENST00000367025.3 |
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr9_+_15422702 | 0.04 |
ENST00000380821.3
ENST00000421710.1 |
SNAPC3
|
small nuclear RNA activating complex, polypeptide 3, 50kDa |
| chr17_+_75315654 | 0.04 |
ENST00000590595.1
|
SEPT9
|
septin 9 |
| chr16_+_85942594 | 0.04 |
ENST00000566369.1
|
IRF8
|
interferon regulatory factor 8 |
| chr11_+_108093755 | 0.04 |
ENST00000527891.1
ENST00000532931.1 |
ATM
|
ataxia telangiectasia mutated |
| chr2_-_18741882 | 0.04 |
ENST00000381249.3
|
RDH14
|
retinol dehydrogenase 14 (all-trans/9-cis/11-cis) |
| chr19_-_8567478 | 0.04 |
ENST00000255612.3
|
PRAM1
|
PML-RARA regulated adaptor molecule 1 |
| chr5_-_94417186 | 0.04 |
ENST00000312216.8
ENST00000512425.1 |
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr6_+_144904334 | 0.04 |
ENST00000367526.4
|
UTRN
|
utrophin |
| chr16_+_28505955 | 0.04 |
ENST00000564831.1
ENST00000328423.5 ENST00000431282.1 |
APOBR
|
apolipoprotein B receptor |
| chr17_-_67057047 | 0.04 |
ENST00000495634.1
ENST00000453985.2 ENST00000585714.1 |
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr5_+_159895275 | 0.04 |
ENST00000517927.1
|
MIR146A
|
microRNA 146a |
| chr1_+_239882842 | 0.04 |
ENST00000448020.1
|
CHRM3
|
cholinergic receptor, muscarinic 3 |
| chr7_+_102389434 | 0.04 |
ENST00000409231.3
ENST00000418198.1 |
FAM185A
|
family with sequence similarity 185, member A |
| chr1_-_206945830 | 0.04 |
ENST00000423557.1
|
IL10
|
interleukin 10 |
| chr3_+_96533413 | 0.04 |
ENST00000470610.2
ENST00000389672.5 |
EPHA6
|
EPH receptor A6 |
| chr4_+_76481258 | 0.04 |
ENST00000311623.4
ENST00000435974.2 |
C4orf26
|
chromosome 4 open reading frame 26 |
| chr6_+_131571535 | 0.04 |
ENST00000474850.2
|
AKAP7
|
A kinase (PRKA) anchor protein 7 |
| chr17_-_41836153 | 0.04 |
ENST00000301691.2
|
SOST
|
sclerostin |
| chr19_+_11466062 | 0.04 |
ENST00000251473.5
ENST00000591329.1 ENST00000586380.1 |
DKFZP761J1410
|
Lipid phosphate phosphatase-related protein type 2 |
| chr3_-_71179988 | 0.04 |
ENST00000491238.1
|
FOXP1
|
forkhead box P1 |
| chr3_-_71179699 | 0.04 |
ENST00000497355.1
|
FOXP1
|
forkhead box P1 |
| chr4_+_89444961 | 0.04 |
ENST00000513325.1
|
HERC3
|
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
| chr3_+_98699880 | 0.03 |
ENST00000473756.1
|
LINC00973
|
long intergenic non-protein coding RNA 973 |
| chr21_-_36421401 | 0.03 |
ENST00000486278.2
|
RUNX1
|
runt-related transcription factor 1 |
| chr5_+_35856951 | 0.03 |
ENST00000303115.3
ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr11_+_121447469 | 0.03 |
ENST00000532694.1
ENST00000534286.1 |
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing |
| chr10_-_29811456 | 0.03 |
ENST00000535393.1
|
SVIL
|
supervillin |
| chr15_+_63335899 | 0.03 |
ENST00000561266.1
|
TPM1
|
tropomyosin 1 (alpha) |
| chr5_+_158737824 | 0.03 |
ENST00000521472.1
|
AC008697.1
|
AC008697.1 |
| chr4_-_6474173 | 0.03 |
ENST00000382599.4
|
PPP2R2C
|
protein phosphatase 2, regulatory subunit B, gamma |
| chr12_-_27090896 | 0.03 |
ENST00000539625.1
ENST00000538727.1 |
ASUN
|
asunder spermatogenesis regulator |
| chr8_-_72274467 | 0.03 |
ENST00000340726.3
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr2_-_4703793 | 0.03 |
ENST00000421212.1
ENST00000412134.1 |
AC022311.1
|
AC022311.1 |
| chr2_+_208423891 | 0.03 |
ENST00000448277.1
ENST00000457101.1 |
CREB1
|
cAMP responsive element binding protein 1 |
| chr3_+_141105705 | 0.03 |
ENST00000513258.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr10_+_112404132 | 0.03 |
ENST00000369519.3
|
RBM20
|
RNA binding motif protein 20 |
| chr21_+_17214724 | 0.03 |
ENST00000449491.1
|
USP25
|
ubiquitin specific peptidase 25 |
| chr17_-_56494908 | 0.03 |
ENST00000577716.1
|
RNF43
|
ring finger protein 43 |
| chr11_+_114310164 | 0.03 |
ENST00000544196.1
ENST00000539754.1 ENST00000539275.1 |
REXO2
|
RNA exonuclease 2 |
| chr1_-_150978953 | 0.03 |
ENST00000493834.2
|
FAM63A
|
family with sequence similarity 63, member A |
| chr1_+_110453514 | 0.03 |
ENST00000369802.3
ENST00000420111.2 |
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr1_-_161277210 | 0.03 |
ENST00000491222.2
|
MPZ
|
myelin protein zero |
| chr2_+_97203082 | 0.03 |
ENST00000454558.2
|
ARID5A
|
AT rich interactive domain 5A (MRF1-like) |
| chr19_-_42636617 | 0.03 |
ENST00000529067.1
ENST00000529952.1 ENST00000533720.1 ENST00000389341.5 ENST00000342301.4 |
POU2F2
|
POU class 2 homeobox 2 |
| chr3_+_156799587 | 0.03 |
ENST00000469196.1
|
RP11-6F2.5
|
RP11-6F2.5 |
| chr5_+_148206156 | 0.03 |
ENST00000305988.4
|
ADRB2
|
adrenoceptor beta 2, surface |
| chr1_-_113161730 | 0.03 |
ENST00000544629.1
ENST00000543570.1 ENST00000360743.4 ENST00000490067.1 ENST00000343210.7 ENST00000369666.1 |
ST7L
|
suppression of tumorigenicity 7 like |
| chr1_+_164528437 | 0.03 |
ENST00000485769.1
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr19_-_51875894 | 0.03 |
ENST00000600427.1
ENST00000595217.1 ENST00000221978.5 |
NKG7
|
natural killer cell group 7 sequence |
| chr3_-_196014520 | 0.03 |
ENST00000441879.1
ENST00000292823.2 ENST00000411591.1 ENST00000431016.1 ENST00000443555.1 |
PCYT1A
|
phosphate cytidylyltransferase 1, choline, alpha |
| chr9_-_37384431 | 0.03 |
ENST00000452923.1
|
RP11-397D12.4
|
RP11-397D12.4 |
| chr21_+_35552978 | 0.03 |
ENST00000428914.2
ENST00000609062.1 ENST00000609947.1 |
LINC00310
|
long intergenic non-protein coding RNA 310 |
| chr17_-_7164410 | 0.03 |
ENST00000574070.1
|
CLDN7
|
claudin 7 |
| chr12_+_108079509 | 0.03 |
ENST00000412830.3
ENST00000547995.1 |
PWP1
|
PWP1 homolog (S. cerevisiae) |
| chr11_+_114310237 | 0.03 |
ENST00000539119.1
|
REXO2
|
RNA exonuclease 2 |
| chr2_-_61389240 | 0.03 |
ENST00000606876.1
|
RP11-493E12.1
|
RP11-493E12.1 |
| chr3_+_187957646 | 0.03 |
ENST00000457242.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr1_-_92951607 | 0.03 |
ENST00000427103.1
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr20_-_25062767 | 0.03 |
ENST00000429762.3
ENST00000444511.2 ENST00000376707.3 |
VSX1
|
visual system homeobox 1 |
| chr15_-_55562451 | 0.03 |
ENST00000568803.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr3_-_98241760 | 0.03 |
ENST00000507874.1
ENST00000502299.1 ENST00000508659.1 ENST00000510545.1 ENST00000511667.1 ENST00000394185.2 ENST00000394181.2 ENST00000508902.1 ENST00000341181.6 ENST00000437922.1 ENST00000394180.2 |
CLDND1
|
claudin domain containing 1 |
| chr4_+_106816592 | 0.03 |
ENST00000379987.2
ENST00000453617.2 ENST00000427316.2 ENST00000514622.1 ENST00000305572.8 |
NPNT
|
nephronectin |
| chr19_+_50338234 | 0.03 |
ENST00000593767.1
|
MED25
|
mediator complex subunit 25 |
| chr1_-_150979333 | 0.03 |
ENST00000312210.5
|
FAM63A
|
family with sequence similarity 63, member A |
| chr6_-_146285455 | 0.03 |
ENST00000367505.2
|
SHPRH
|
SNF2 histone linker PHD RING helicase, E3 ubiquitin protein ligase |
| chr9_-_123342415 | 0.03 |
ENST00000349780.4
ENST00000360190.4 ENST00000360822.3 ENST00000359309.3 |
CDK5RAP2
|
CDK5 regulatory subunit associated protein 2 |
| chr11_-_58345569 | 0.03 |
ENST00000528954.1
ENST00000528489.1 |
LPXN
|
leupaxin |
| chr17_-_56494882 | 0.03 |
ENST00000584437.1
|
RNF43
|
ring finger protein 43 |
| chr2_-_74555350 | 0.03 |
ENST00000444570.1
|
SLC4A5
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 5 |
| chr1_+_221051699 | 0.03 |
ENST00000366903.6
|
HLX
|
H2.0-like homeobox |
| chr8_-_60031762 | 0.03 |
ENST00000361421.1
|
TOX
|
thymocyte selection-associated high mobility group box |
| chr6_+_63921399 | 0.03 |
ENST00000356170.3
|
FKBP1C
|
FK506 binding protein 1C |
| chr3_-_72897545 | 0.03 |
ENST00000325599.8
|
SHQ1
|
SHQ1, H/ACA ribonucleoprotein assembly factor |
| chrX_-_106146547 | 0.03 |
ENST00000276173.4
ENST00000411805.1 |
RIPPLY1
|
ripply transcriptional repressor 1 |
| chr2_+_169659121 | 0.03 |
ENST00000397206.2
ENST00000397209.2 ENST00000421711.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr6_-_146285221 | 0.03 |
ENST00000367503.3
ENST00000438092.2 ENST00000275233.7 |
SHPRH
|
SNF2 histone linker PHD RING helicase, E3 ubiquitin protein ligase |
| chr5_+_172386517 | 0.03 |
ENST00000519522.1
|
RPL26L1
|
ribosomal protein L26-like 1 |
| chr11_-_108093329 | 0.03 |
ENST00000278612.8
|
NPAT
|
nuclear protein, ataxia-telangiectasia locus |
| chr11_+_114310102 | 0.03 |
ENST00000265881.5
|
REXO2
|
RNA exonuclease 2 |
| chr1_+_150039877 | 0.03 |
ENST00000419023.1
|
VPS45
|
vacuolar protein sorting 45 homolog (S. cerevisiae) |
| chr2_-_175462456 | 0.03 |
ENST00000409891.1
ENST00000410117.1 |
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr12_-_49245916 | 0.03 |
ENST00000552512.1
ENST00000551468.1 |
DDX23
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 23 |
| chr8_-_99955042 | 0.03 |
ENST00000519420.1
|
STK3
|
serine/threonine kinase 3 |
| chr12_-_323689 | 0.03 |
ENST00000428720.1
|
SLC6A12
|
solute carrier family 6 (neurotransmitter transporter), member 12 |
| chr3_+_141105235 | 0.03 |
ENST00000503809.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr8_+_91233711 | 0.03 |
ENST00000523283.1
ENST00000517400.1 |
LINC00534
|
long intergenic non-protein coding RNA 534 |
| chr4_+_78783674 | 0.03 |
ENST00000315567.8
|
MRPL1
|
mitochondrial ribosomal protein L1 |
| chr3_-_49170405 | 0.03 |
ENST00000305544.4
ENST00000494831.1 |
LAMB2
|
laminin, beta 2 (laminin S) |
| chr2_+_30455016 | 0.03 |
ENST00000401506.1
ENST00000407930.2 |
LBH
|
limb bud and heart development |
| chr3_+_96533621 | 0.03 |
ENST00000542517.1
ENST00000506569.1 |
EPHA6
|
EPH receptor A6 |
| chr10_+_115511434 | 0.03 |
ENST00000369312.4
|
PLEKHS1
|
pleckstrin homology domain containing, family S member 1 |
| chr16_+_29674540 | 0.03 |
ENST00000436527.1
ENST00000360121.3 ENST00000449759.1 |
SPN
QPRT
|
sialophorin quinolinate phosphoribosyltransferase |
| chr12_+_54410664 | 0.02 |
ENST00000303406.4
|
HOXC4
|
homeobox C4 |
| chr17_-_56082455 | 0.02 |
ENST00000578794.1
|
RP11-159D12.5
|
Uncharacterized protein |
| chr6_+_143771934 | 0.02 |
ENST00000367592.1
|
PEX3
|
peroxisomal biogenesis factor 3 |
| chr1_-_86043921 | 0.02 |
ENST00000535924.2
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr11_+_35198118 | 0.02 |
ENST00000525211.1
ENST00000526000.1 ENST00000279452.6 ENST00000527889.1 |
CD44
|
CD44 molecule (Indian blood group) |
| chr5_-_137878887 | 0.02 |
ENST00000507939.1
ENST00000572514.1 ENST00000499810.2 ENST00000360541.5 |
ETF1
|
eukaryotic translation termination factor 1 |
| chr4_+_184020398 | 0.02 |
ENST00000403733.3
ENST00000378925.3 |
WWC2
|
WW and C2 domain containing 2 |
| chr5_+_145316120 | 0.02 |
ENST00000359120.4
|
SH3RF2
|
SH3 domain containing ring finger 2 |
| chr1_+_152486950 | 0.02 |
ENST00000368790.3
|
CRCT1
|
cysteine-rich C-terminal 1 |
| chr19_+_13135731 | 0.02 |
ENST00000587260.1
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr17_-_44270133 | 0.02 |
ENST00000262419.6
ENST00000393476.3 |
KANSL1
|
KAT8 regulatory NSL complex subunit 1 |
| chr8_-_101718991 | 0.02 |
ENST00000517990.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr4_+_88896819 | 0.02 |
ENST00000237623.7
ENST00000395080.3 ENST00000508233.1 ENST00000360804.4 |
SPP1
|
secreted phosphoprotein 1 |
| chr1_+_110453608 | 0.02 |
ENST00000369801.1
|
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr1_+_150039787 | 0.02 |
ENST00000535106.1
|
VPS45
|
vacuolar protein sorting 45 homolog (S. cerevisiae) |
| chr2_-_198299726 | 0.02 |
ENST00000409915.4
ENST00000487698.1 ENST00000414963.2 ENST00000335508.6 |
SF3B1
|
splicing factor 3b, subunit 1, 155kDa |
| chr11_-_118436707 | 0.02 |
ENST00000264020.2
ENST00000264021.3 |
IFT46
|
intraflagellar transport 46 homolog (Chlamydomonas) |
| chr6_-_42418999 | 0.02 |
ENST00000340840.2
ENST00000354325.2 |
TRERF1
|
transcriptional regulating factor 1 |
| chr5_-_159546396 | 0.02 |
ENST00000523662.1
ENST00000456329.3 ENST00000307063.7 |
PWWP2A
|
PWWP domain containing 2A |
| chr10_-_65028817 | 0.02 |
ENST00000542921.1
|
JMJD1C
|
jumonji domain containing 1C |
| chr15_+_67390920 | 0.02 |
ENST00000559092.1
ENST00000560175.1 |
SMAD3
|
SMAD family member 3 |
| chr17_+_38673270 | 0.02 |
ENST00000578280.1
|
RP5-1028K7.2
|
RP5-1028K7.2 |
| chr12_+_19358228 | 0.02 |
ENST00000424268.1
ENST00000543806.1 |
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of RUNX3_BCL11A
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0060611 | mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) positive regulation of microglial cell migration(GO:1904141) |
| 0.0 | 0.1 | GO:0032714 | regulation of tolerance induction dependent upon immune response(GO:0002652) positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) negative regulation of interleukin-5 production(GO:0032714) positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.0 | 0.1 | GO:0033594 | response to vitamin K(GO:0032571) response to hydroxyisoflavone(GO:0033594) |
| 0.0 | 0.2 | GO:0071104 | response to interleukin-9(GO:0071104) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.1 | GO:0002625 | regulation of T cell antigen processing and presentation(GO:0002625) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.0 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.0 | 0.2 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.0 | GO:1902955 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.0 | 0.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.0 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.0 | 0.0 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.1 | GO:0045423 | regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) |
| 0.0 | 0.0 | GO:0060437 | lung growth(GO:0060437) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.0 | 0.1 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.0 | 0.0 | GO:0032279 | asymmetric synapse(GO:0032279) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.0 | 0.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.1 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.0 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.0 | 0.0 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.0 | 0.0 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.0 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |