Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
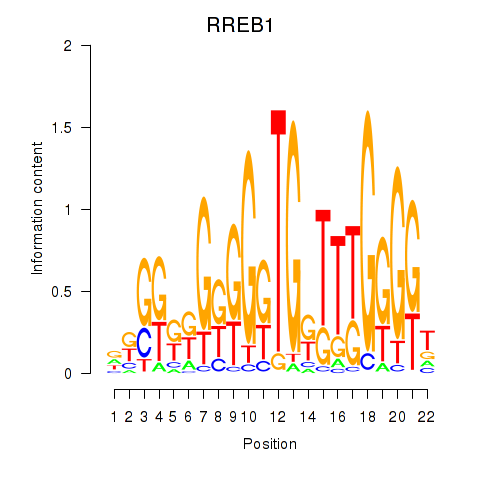
Results for RREB1
Z-value: 0.96
Transcription factors associated with RREB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RREB1
|
ENSG00000124782.15 | ras responsive element binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RREB1 | hg19_v2_chr6_+_7107999_7108054 | -0.23 | 7.7e-01 | Click! |
Activity profile of RREB1 motif
Sorted Z-values of RREB1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_1093318 | 0.88 |
ENST00000333592.6
|
MUC2
|
mucin 2, oligomeric mucus/gel-forming |
| chr13_-_74708372 | 0.52 |
ENST00000377666.4
|
KLF12
|
Kruppel-like factor 12 |
| chr8_+_26149274 | 0.52 |
ENST00000522535.1
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha |
| chr15_+_96897466 | 0.50 |
ENST00000558382.1
ENST00000558499.1 |
RP11-522B15.3
|
RP11-522B15.3 |
| chr17_-_58499766 | 0.45 |
ENST00000588898.1
|
USP32
|
ubiquitin specific peptidase 32 |
| chrX_+_17755696 | 0.43 |
ENST00000419185.1
|
SCML1
|
sex comb on midleg-like 1 (Drosophila) |
| chr19_-_30205963 | 0.43 |
ENST00000392278.2
|
C19orf12
|
chromosome 19 open reading frame 12 |
| chr7_+_91570240 | 0.42 |
ENST00000394564.1
|
AKAP9
|
A kinase (PRKA) anchor protein 9 |
| chr12_-_62653903 | 0.38 |
ENST00000552075.1
|
FAM19A2
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2 |
| chr4_-_56412713 | 0.37 |
ENST00000435527.2
|
CLOCK
|
clock circadian regulator |
| chr12_+_133033703 | 0.36 |
ENST00000542627.1
|
RP11-503G7.1
|
RP11-503G7.1 |
| chr12_+_96588279 | 0.35 |
ENST00000552142.1
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr4_-_100484825 | 0.34 |
ENST00000273962.3
ENST00000514547.1 ENST00000455368.2 |
TRMT10A
|
tRNA methyltransferase 10 homolog A (S. cerevisiae) |
| chr17_+_15604513 | 0.32 |
ENST00000481540.1
|
ZNF286A
|
Homo sapiens zinc finger protein 286A (ZNF286A), transcript variant 6, mRNA. |
| chr11_-_61196858 | 0.31 |
ENST00000413184.2
|
CPSF7
|
cleavage and polyadenylation specific factor 7, 59kDa |
| chr19_+_42381173 | 0.31 |
ENST00000221972.3
|
CD79A
|
CD79a molecule, immunoglobulin-associated alpha |
| chr11_-_96239990 | 0.30 |
ENST00000511243.2
|
JRKL-AS1
|
JRKL antisense RNA 1 |
| chr13_-_52026730 | 0.30 |
ENST00000420668.2
|
INTS6
|
integrator complex subunit 6 |
| chr19_+_42381337 | 0.29 |
ENST00000597454.1
ENST00000444740.2 |
CD79A
|
CD79a molecule, immunoglobulin-associated alpha |
| chr19_+_2164173 | 0.29 |
ENST00000452696.1
|
DOT1L
|
DOT1-like histone H3K79 methyltransferase |
| chr10_+_115438920 | 0.28 |
ENST00000429617.1
ENST00000369331.4 |
CASP7
|
caspase 7, apoptosis-related cysteine peptidase |
| chrX_+_17755563 | 0.27 |
ENST00000380045.3
ENST00000380041.3 ENST00000380043.3 ENST00000398080.1 |
SCML1
|
sex comb on midleg-like 1 (Drosophila) |
| chr2_-_74645669 | 0.26 |
ENST00000518401.1
|
C2orf81
|
chromosome 2 open reading frame 81 |
| chr13_-_52027134 | 0.26 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr11_-_129872672 | 0.24 |
ENST00000531431.1
ENST00000527581.1 |
PRDM10
|
PR domain containing 10 |
| chr10_-_112064665 | 0.24 |
ENST00000369603.5
|
SMNDC1
|
survival motor neuron domain containing 1 |
| chr7_-_92465868 | 0.23 |
ENST00000424848.2
|
CDK6
|
cyclin-dependent kinase 6 |
| chr15_+_63481668 | 0.23 |
ENST00000321437.4
ENST00000559006.1 ENST00000448330.2 |
RAB8B
|
RAB8B, member RAS oncogene family |
| chr3_+_141205852 | 0.23 |
ENST00000286364.3
ENST00000452898.1 |
RASA2
|
RAS p21 protein activator 2 |
| chr4_-_100485183 | 0.23 |
ENST00000394876.2
|
TRMT10A
|
tRNA methyltransferase 10 homolog A (S. cerevisiae) |
| chr17_-_4642429 | 0.23 |
ENST00000573123.1
|
CXCL16
|
chemokine (C-X-C motif) ligand 16 |
| chr13_+_111972980 | 0.23 |
ENST00000283547.1
|
TEX29
|
testis expressed 29 |
| chrX_+_102631844 | 0.22 |
ENST00000372634.1
ENST00000299872.7 |
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1 |
| chr3_+_187461442 | 0.22 |
ENST00000450760.1
|
RP11-211G3.2
|
RP11-211G3.2 |
| chr7_+_91570165 | 0.22 |
ENST00000356239.3
ENST00000359028.2 ENST00000358100.2 |
AKAP9
|
A kinase (PRKA) anchor protein 9 |
| chr3_-_71632894 | 0.21 |
ENST00000493089.1
|
FOXP1
|
forkhead box P1 |
| chr19_+_36157715 | 0.21 |
ENST00000379013.2
ENST00000222275.2 |
UPK1A
|
uroplakin 1A |
| chrX_+_151867214 | 0.20 |
ENST00000329342.5
ENST00000412733.1 ENST00000457643.1 |
MAGEA6
|
melanoma antigen family A, 6 |
| chr16_+_14396121 | 0.20 |
ENST00000570945.1
|
RP11-65J21.3
|
RP11-65J21.3 |
| chr16_-_4588469 | 0.20 |
ENST00000588381.1
ENST00000563332.2 |
CDIP1
|
cell death-inducing p53 target 1 |
| chr6_-_86352982 | 0.20 |
ENST00000369622.3
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr18_+_56531584 | 0.19 |
ENST00000590287.1
|
ZNF532
|
zinc finger protein 532 |
| chr7_-_154794763 | 0.19 |
ENST00000404141.1
|
PAXIP1
|
PAX interacting (with transcription-activation domain) protein 1 |
| chr19_-_51071302 | 0.19 |
ENST00000389201.3
ENST00000600381.1 |
LRRC4B
|
leucine rich repeat containing 4B |
| chr7_-_154794621 | 0.19 |
ENST00000419436.1
ENST00000397192.1 |
PAXIP1
|
PAX interacting (with transcription-activation domain) protein 1 |
| chr22_-_37976082 | 0.19 |
ENST00000215886.4
|
LGALS2
|
lectin, galactoside-binding, soluble, 2 |
| chr6_-_86353510 | 0.18 |
ENST00000444272.1
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr18_-_45456693 | 0.18 |
ENST00000587421.1
|
SMAD2
|
SMAD family member 2 |
| chr15_+_77713299 | 0.18 |
ENST00000559099.1
|
HMG20A
|
high mobility group 20A |
| chr14_+_91709279 | 0.18 |
ENST00000554096.1
|
CTD-2547L24.3
|
HCG1816139; Uncharacterized protein |
| chr19_-_3029011 | 0.18 |
ENST00000590536.1
ENST00000587137.1 ENST00000455444.2 ENST00000262953.6 |
TLE2
|
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila) |
| chr17_-_76732928 | 0.17 |
ENST00000589768.1
|
CYTH1
|
cytohesin 1 |
| chr2_+_46926326 | 0.17 |
ENST00000394861.2
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr17_-_7082668 | 0.17 |
ENST00000573083.1
ENST00000574388.1 |
ASGR1
|
asialoglycoprotein receptor 1 |
| chr12_+_44152740 | 0.17 |
ENST00000440781.2
ENST00000431837.1 ENST00000550616.1 ENST00000448290.2 ENST00000551736.1 |
IRAK4
|
interleukin-1 receptor-associated kinase 4 |
| chr19_-_47128294 | 0.17 |
ENST00000596260.1
ENST00000597185.1 ENST00000598865.1 ENST00000594275.1 ENST00000291294.2 |
PTGIR
|
prostaglandin I2 (prostacyclin) receptor (IP) |
| chr17_+_41177220 | 0.17 |
ENST00000587250.2
ENST00000544533.1 |
RND2
|
Rho family GTPase 2 |
| chr10_+_13628933 | 0.16 |
ENST00000417658.1
ENST00000320054.4 |
PRPF18
|
pre-mRNA processing factor 18 |
| chr19_+_55795493 | 0.16 |
ENST00000309383.1
|
BRSK1
|
BR serine/threonine kinase 1 |
| chr8_-_66753682 | 0.16 |
ENST00000396642.3
|
PDE7A
|
phosphodiesterase 7A |
| chr18_-_21242774 | 0.16 |
ENST00000322980.9
|
ANKRD29
|
ankyrin repeat domain 29 |
| chr1_-_113392399 | 0.16 |
ENST00000449572.2
ENST00000433505.1 |
RP11-426L16.8
|
RP11-426L16.8 |
| chr19_-_3025614 | 0.16 |
ENST00000447365.2
|
TLE2
|
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila) |
| chr17_-_62658186 | 0.16 |
ENST00000262435.9
|
SMURF2
|
SMAD specific E3 ubiquitin protein ligase 2 |
| chr8_+_32406179 | 0.16 |
ENST00000405005.3
|
NRG1
|
neuregulin 1 |
| chrX_-_14891150 | 0.16 |
ENST00000452869.1
ENST00000398334.1 ENST00000324138.3 |
FANCB
|
Fanconi anemia, complementation group B |
| chr17_+_30348024 | 0.16 |
ENST00000327564.7
ENST00000584368.1 ENST00000394713.3 ENST00000341671.7 |
LRRC37B
|
leucine rich repeat containing 37B |
| chr3_-_52869205 | 0.16 |
ENST00000446157.2
|
MUSTN1
|
musculoskeletal, embryonic nuclear protein 1 |
| chr6_+_42749759 | 0.15 |
ENST00000314073.5
|
GLTSCR1L
|
GLTSCR1-like |
| chrX_-_131352152 | 0.15 |
ENST00000342983.2
|
RAP2C
|
RAP2C, member of RAS oncogene family |
| chr10_+_70091847 | 0.15 |
ENST00000441000.2
ENST00000354695.5 |
HNRNPH3
|
heterogeneous nuclear ribonucleoprotein H3 (2H9) |
| chr20_-_58508359 | 0.15 |
ENST00000446834.1
|
SYCP2
|
synaptonemal complex protein 2 |
| chr18_-_21242729 | 0.15 |
ENST00000585908.2
|
ANKRD29
|
ankyrin repeat domain 29 |
| chr20_+_33462745 | 0.15 |
ENST00000488172.1
|
ACSS2
|
acyl-CoA synthetase short-chain family member 2 |
| chr22_+_32750872 | 0.15 |
ENST00000397468.1
|
RFPL3
|
ret finger protein-like 3 |
| chr12_-_56101647 | 0.14 |
ENST00000347027.6
ENST00000257879.6 ENST00000257880.7 ENST00000394230.2 ENST00000394229.2 |
ITGA7
|
integrin, alpha 7 |
| chr22_-_44258280 | 0.14 |
ENST00000540422.1
|
SULT4A1
|
sulfotransferase family 4A, member 1 |
| chr4_+_190992087 | 0.14 |
ENST00000553598.1
|
DUX4L7
|
double homeobox 4 like 7 |
| chr16_+_28889801 | 0.14 |
ENST00000395503.4
|
ATP2A1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr3_+_169684553 | 0.14 |
ENST00000337002.4
ENST00000480708.1 |
SEC62
|
SEC62 homolog (S. cerevisiae) |
| chr12_+_57522692 | 0.14 |
ENST00000554174.1
|
LRP1
|
low density lipoprotein receptor-related protein 1 |
| chr12_-_44152551 | 0.14 |
ENST00000416848.2
ENST00000550784.1 ENST00000547156.1 ENST00000549868.1 ENST00000553166.1 ENST00000551923.1 ENST00000431332.3 ENST00000344862.5 |
PUS7L
|
pseudouridylate synthase 7 homolog (S. cerevisiae)-like |
| chr18_+_9334755 | 0.14 |
ENST00000262120.5
|
TWSG1
|
twisted gastrulation BMP signaling modulator 1 |
| chr5_+_78908388 | 0.14 |
ENST00000296783.3
|
PAPD4
|
PAP associated domain containing 4 |
| chr9_-_13279563 | 0.13 |
ENST00000541718.1
|
MPDZ
|
multiple PDZ domain protein |
| chr16_+_57680043 | 0.13 |
ENST00000569154.1
|
GPR56
|
G protein-coupled receptor 56 |
| chr9_-_13279406 | 0.13 |
ENST00000546205.1
|
MPDZ
|
multiple PDZ domain protein |
| chr12_-_131323719 | 0.13 |
ENST00000392373.2
|
STX2
|
syntaxin 2 |
| chr15_-_89764929 | 0.13 |
ENST00000268125.5
|
RLBP1
|
retinaldehyde binding protein 1 |
| chr11_+_34073269 | 0.13 |
ENST00000389645.3
|
CAPRIN1
|
cell cycle associated protein 1 |
| chr6_+_151646800 | 0.13 |
ENST00000354675.6
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr2_-_230786378 | 0.13 |
ENST00000430954.1
|
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr3_+_140660743 | 0.13 |
ENST00000453248.2
|
SLC25A36
|
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36 |
| chr19_-_44084586 | 0.13 |
ENST00000599693.1
ENST00000598165.1 |
XRCC1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr2_-_54087066 | 0.13 |
ENST00000394705.2
ENST00000352846.3 ENST00000406625.2 |
GPR75
GPR75-ASB3
ASB3
|
G protein-coupled receptor 75 GPR75-ASB3 readthrough Ankyrin repeat and SOCS box protein 3 |
| chr3_+_154797781 | 0.13 |
ENST00000382989.3
|
MME
|
membrane metallo-endopeptidase |
| chr4_-_13549417 | 0.13 |
ENST00000501050.1
|
AC006445.8
|
long intergenic non-protein coding RNA 1096 |
| chr4_-_100485143 | 0.13 |
ENST00000394877.3
|
TRMT10A
|
tRNA methyltransferase 10 homolog A (S. cerevisiae) |
| chr9_-_13279589 | 0.13 |
ENST00000319217.7
|
MPDZ
|
multiple PDZ domain protein |
| chrX_-_40036520 | 0.12 |
ENST00000406200.2
ENST00000378455.4 ENST00000342274.4 |
BCOR
|
BCL6 corepressor |
| chr16_+_68771128 | 0.12 |
ENST00000261769.5
ENST00000422392.2 |
CDH1
|
cadherin 1, type 1, E-cadherin (epithelial) |
| chr4_+_75230853 | 0.12 |
ENST00000244869.2
|
EREG
|
epiregulin |
| chr1_+_43148625 | 0.12 |
ENST00000436427.1
|
YBX1
|
Y box binding protein 1 |
| chr17_-_39677971 | 0.12 |
ENST00000393976.2
|
KRT15
|
keratin 15 |
| chr11_-_61197187 | 0.12 |
ENST00000449811.1
ENST00000413232.1 ENST00000340437.4 ENST00000539952.1 ENST00000544585.1 ENST00000450000.1 |
CPSF7
|
cleavage and polyadenylation specific factor 7, 59kDa |
| chr5_-_98262240 | 0.12 |
ENST00000284049.3
|
CHD1
|
chromodomain helicase DNA binding protein 1 |
| chr4_-_6474173 | 0.12 |
ENST00000382599.4
|
PPP2R2C
|
protein phosphatase 2, regulatory subunit B, gamma |
| chr11_-_61197406 | 0.12 |
ENST00000541963.1
ENST00000477890.2 |
CPSF7
|
cleavage and polyadenylation specific factor 7, 59kDa |
| chr11_-_22881972 | 0.12 |
ENST00000532798.2
|
CCDC179
|
coiled-coil domain containing 179 |
| chr7_+_97840739 | 0.12 |
ENST00000609256.1
|
BHLHA15
|
basic helix-loop-helix family, member a15 |
| chr20_+_1115821 | 0.12 |
ENST00000435720.1
|
PSMF1
|
proteasome (prosome, macropain) inhibitor subunit 1 (PI31) |
| chr7_+_152456829 | 0.11 |
ENST00000377776.3
ENST00000256001.8 ENST00000397282.2 |
ACTR3B
|
ARP3 actin-related protein 3 homolog B (yeast) |
| chr12_-_123849374 | 0.11 |
ENST00000602398.1
ENST00000602750.1 |
SBNO1
|
strawberry notch homolog 1 (Drosophila) |
| chr21_+_44073860 | 0.11 |
ENST00000335512.4
ENST00000539837.1 ENST00000291539.6 ENST00000380328.2 ENST00000398232.3 ENST00000398234.3 ENST00000398236.3 ENST00000328862.6 ENST00000335440.6 ENST00000398225.3 ENST00000398229.3 ENST00000398227.3 |
PDE9A
|
phosphodiesterase 9A |
| chr5_-_142065612 | 0.11 |
ENST00000360966.5
ENST00000411960.1 |
FGF1
|
fibroblast growth factor 1 (acidic) |
| chr11_+_17756279 | 0.11 |
ENST00000265969.6
|
KCNC1
|
potassium voltage-gated channel, Shaw-related subfamily, member 1 |
| chr17_-_66453562 | 0.11 |
ENST00000262139.5
ENST00000546360.1 |
WIPI1
|
WD repeat domain, phosphoinositide interacting 1 |
| chr13_-_77601282 | 0.11 |
ENST00000355619.5
|
FBXL3
|
F-box and leucine-rich repeat protein 3 |
| chr10_+_70091812 | 0.11 |
ENST00000265866.7
|
HNRNPH3
|
heterogeneous nuclear ribonucleoprotein H3 (2H9) |
| chr12_+_123849462 | 0.11 |
ENST00000543072.1
|
hsa-mir-8072
|
hsa-mir-8072 |
| chr12_-_16760195 | 0.11 |
ENST00000546281.1
ENST00000537757.1 |
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr16_+_50280020 | 0.11 |
ENST00000564965.1
|
ADCY7
|
adenylate cyclase 7 |
| chr15_+_77712993 | 0.10 |
ENST00000336216.4
ENST00000381714.3 ENST00000558651.1 |
HMG20A
|
high mobility group 20A |
| chr6_+_7541808 | 0.10 |
ENST00000379802.3
|
DSP
|
desmoplakin |
| chr13_+_24734880 | 0.10 |
ENST00000382095.4
|
SPATA13
|
spermatogenesis associated 13 |
| chr7_+_114562172 | 0.10 |
ENST00000393486.1
ENST00000257724.3 |
MDFIC
|
MyoD family inhibitor domain containing |
| chr11_+_61197572 | 0.10 |
ENST00000542074.1
ENST00000534878.1 ENST00000537782.1 ENST00000543265.1 |
SDHAF2
|
succinate dehydrogenase complex assembly factor 2 |
| chr13_+_60971427 | 0.10 |
ENST00000535286.1
ENST00000377881.2 |
TDRD3
|
tudor domain containing 3 |
| chr3_-_116163830 | 0.10 |
ENST00000333617.4
|
LSAMP
|
limbic system-associated membrane protein |
| chr5_+_133477905 | 0.10 |
ENST00000517799.1
|
TCF7
|
transcription factor 7 (T-cell specific, HMG-box) |
| chr21_-_34915123 | 0.10 |
ENST00000438059.1
|
GART
|
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
| chr2_+_128377550 | 0.10 |
ENST00000437387.1
ENST00000409090.1 |
MYO7B
|
myosin VIIB |
| chr3_+_140660634 | 0.10 |
ENST00000446041.2
ENST00000507429.1 ENST00000324194.6 |
SLC25A36
|
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36 |
| chr12_-_12715266 | 0.10 |
ENST00000228862.2
|
DUSP16
|
dual specificity phosphatase 16 |
| chr11_-_8739383 | 0.10 |
ENST00000531060.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr1_+_110091189 | 0.10 |
ENST00000369851.4
|
GNAI3
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 |
| chr21_+_44073916 | 0.10 |
ENST00000349112.3
ENST00000398224.3 |
PDE9A
|
phosphodiesterase 9A |
| chr8_+_90914859 | 0.10 |
ENST00000520659.1
|
OSGIN2
|
oxidative stress induced growth inhibitor family member 2 |
| chr19_-_15442701 | 0.09 |
ENST00000594841.1
ENST00000601941.1 |
BRD4
|
bromodomain containing 4 |
| chrX_+_149737046 | 0.09 |
ENST00000370396.2
ENST00000542741.1 ENST00000543350.1 ENST00000424519.1 ENST00000413012.2 |
MTM1
|
myotubularin 1 |
| chr12_+_62654155 | 0.09 |
ENST00000312635.6
ENST00000393654.3 ENST00000549237.1 |
USP15
|
ubiquitin specific peptidase 15 |
| chr14_+_57735614 | 0.09 |
ENST00000261558.3
|
AP5M1
|
adaptor-related protein complex 5, mu 1 subunit |
| chr1_+_43148059 | 0.09 |
ENST00000321358.7
ENST00000332220.6 |
YBX1
|
Y box binding protein 1 |
| chr2_+_106468204 | 0.09 |
ENST00000425756.1
ENST00000393349.2 |
NCK2
|
NCK adaptor protein 2 |
| chr1_+_212782012 | 0.09 |
ENST00000341491.4
ENST00000366985.1 |
ATF3
|
activating transcription factor 3 |
| chr19_+_55999916 | 0.09 |
ENST00000587166.1
ENST00000389623.6 |
SSC5D
|
scavenger receptor cysteine rich domain containing (5 domains) |
| chr8_+_28748099 | 0.09 |
ENST00000519047.1
|
HMBOX1
|
homeobox containing 1 |
| chr12_+_861717 | 0.09 |
ENST00000535572.1
|
WNK1
|
WNK lysine deficient protein kinase 1 |
| chr8_-_60031762 | 0.09 |
ENST00000361421.1
|
TOX
|
thymocyte selection-associated high mobility group box |
| chr10_-_35379241 | 0.09 |
ENST00000374748.1
ENST00000374749.3 |
CUL2
|
cullin 2 |
| chrX_-_151307020 | 0.09 |
ENST00000370323.4
ENST00000244096.3 ENST00000427322.2 |
MAGEA10
|
melanoma antigen family A, 10 |
| chr17_+_42385927 | 0.09 |
ENST00000426726.3
ENST00000590941.1 ENST00000225441.7 |
RUNDC3A
|
RUN domain containing 3A |
| chr17_-_31620006 | 0.09 |
ENST00000225823.2
|
ASIC2
|
acid-sensing (proton-gated) ion channel 2 |
| chr16_-_49890016 | 0.09 |
ENST00000563137.2
|
ZNF423
|
zinc finger protein 423 |
| chr16_-_3285144 | 0.09 |
ENST00000431561.3
ENST00000396870.4 |
ZNF200
|
zinc finger protein 200 |
| chr2_+_233385173 | 0.09 |
ENST00000449534.2
|
PRSS56
|
protease, serine, 56 |
| chr11_-_74109422 | 0.09 |
ENST00000298198.4
|
PGM2L1
|
phosphoglucomutase 2-like 1 |
| chr2_-_32236002 | 0.09 |
ENST00000404530.1
|
MEMO1
|
mediator of cell motility 1 |
| chr12_-_131323754 | 0.08 |
ENST00000261653.6
|
STX2
|
syntaxin 2 |
| chr19_+_52074502 | 0.08 |
ENST00000545217.1
ENST00000262259.2 ENST00000596504.1 |
ZNF175
|
zinc finger protein 175 |
| chr17_+_1733251 | 0.08 |
ENST00000570451.1
|
RPA1
|
replication protein A1, 70kDa |
| chr1_-_28559502 | 0.08 |
ENST00000263697.4
|
DNAJC8
|
DnaJ (Hsp40) homolog, subfamily C, member 8 |
| chr1_-_150944411 | 0.08 |
ENST00000368949.4
|
CERS2
|
ceramide synthase 2 |
| chr16_-_89008211 | 0.08 |
ENST00000569464.1
ENST00000569443.1 |
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3 |
| chr11_+_111896320 | 0.08 |
ENST00000531306.1
ENST00000537636.1 |
DLAT
|
dihydrolipoamide S-acetyltransferase |
| chr11_+_66406088 | 0.08 |
ENST00000310092.7
ENST00000396053.4 ENST00000408993.2 |
RBM4
|
RNA binding motif protein 4 |
| chr7_-_150721570 | 0.08 |
ENST00000377974.2
ENST00000444312.1 ENST00000605938.1 ENST00000605952.1 |
ATG9B
|
autophagy related 9B |
| chr7_+_75932863 | 0.08 |
ENST00000429938.1
|
HSPB1
|
heat shock 27kDa protein 1 |
| chrX_+_151903253 | 0.08 |
ENST00000452779.2
ENST00000370291.2 |
CSAG1
|
chondrosarcoma associated gene 1 |
| chr11_+_1897697 | 0.08 |
ENST00000432093.1
|
LSP1
|
lymphocyte-specific protein 1 |
| chr8_+_32406137 | 0.08 |
ENST00000521670.1
|
NRG1
|
neuregulin 1 |
| chr17_+_7788104 | 0.08 |
ENST00000380358.4
|
CHD3
|
chromodomain helicase DNA binding protein 3 |
| chr1_-_31712401 | 0.08 |
ENST00000373736.2
|
NKAIN1
|
Na+/K+ transporting ATPase interacting 1 |
| chr17_-_6459802 | 0.08 |
ENST00000262483.8
|
PITPNM3
|
PITPNM family member 3 |
| chr21_-_36259445 | 0.08 |
ENST00000399240.1
|
RUNX1
|
runt-related transcription factor 1 |
| chr12_-_53297432 | 0.08 |
ENST00000546900.1
|
KRT8
|
keratin 8 |
| chr2_+_192543153 | 0.08 |
ENST00000425611.2
|
NABP1
|
nucleic acid binding protein 1 |
| chr11_-_59436453 | 0.08 |
ENST00000300146.9
|
PATL1
|
protein associated with topoisomerase II homolog 1 (yeast) |
| chr8_+_90914757 | 0.08 |
ENST00000451899.2
|
OSGIN2
|
oxidative stress induced growth inhibitor family member 2 |
| chr10_-_126849588 | 0.08 |
ENST00000411419.2
|
CTBP2
|
C-terminal binding protein 2 |
| chr9_-_140335789 | 0.08 |
ENST00000344119.2
ENST00000371506.2 |
ENTPD8
|
ectonucleoside triphosphate diphosphohydrolase 8 |
| chr11_+_7506713 | 0.07 |
ENST00000329293.3
ENST00000534244.1 |
OLFML1
|
olfactomedin-like 1 |
| chr7_-_51384451 | 0.07 |
ENST00000441453.1
ENST00000265136.7 ENST00000395542.2 ENST00000395540.2 |
COBL
|
cordon-bleu WH2 repeat protein |
| chr19_-_12405689 | 0.07 |
ENST00000355684.5
|
ZNF44
|
zinc finger protein 44 |
| chr11_-_118436606 | 0.07 |
ENST00000530872.1
|
IFT46
|
intraflagellar transport 46 homolog (Chlamydomonas) |
| chr11_+_34073195 | 0.07 |
ENST00000341394.4
|
CAPRIN1
|
cell cycle associated protein 1 |
| chr9_+_35906176 | 0.07 |
ENST00000354323.2
|
HRCT1
|
histidine rich carboxyl terminus 1 |
| chr10_-_95360983 | 0.07 |
ENST00000371464.3
|
RBP4
|
retinol binding protein 4, plasma |
| chr12_-_8088871 | 0.07 |
ENST00000075120.7
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr2_-_179343226 | 0.07 |
ENST00000434643.2
|
FKBP7
|
FK506 binding protein 7 |
| chr4_+_123747834 | 0.07 |
ENST00000264498.3
|
FGF2
|
fibroblast growth factor 2 (basic) |
| chr17_+_1733276 | 0.07 |
ENST00000254719.5
|
RPA1
|
replication protein A1, 70kDa |
| chr2_+_234104079 | 0.07 |
ENST00000417661.1
|
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa |
| chr10_+_13628921 | 0.07 |
ENST00000378572.3
|
PRPF18
|
pre-mRNA processing factor 18 |
| chr17_-_73840614 | 0.07 |
ENST00000586108.1
|
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr17_+_55334364 | 0.07 |
ENST00000322684.3
ENST00000579590.1 |
MSI2
|
musashi RNA-binding protein 2 |
| chr16_-_21170762 | 0.07 |
ENST00000261383.3
ENST00000415178.1 |
DNAH3
|
dynein, axonemal, heavy chain 3 |
| chr2_+_223289208 | 0.07 |
ENST00000321276.7
|
SGPP2
|
sphingosine-1-phosphate phosphatase 2 |
| chrX_+_151903207 | 0.07 |
ENST00000370287.3
|
CSAG1
|
chondrosarcoma associated gene 1 |
| chr11_+_111896090 | 0.07 |
ENST00000393051.1
|
DLAT
|
dihydrolipoamide S-acetyltransferase |
Network of associatons between targets according to the STRING database.
First level regulatory network of RREB1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.1 | 0.2 | GO:0071030 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.1 | 0.2 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.1 | 0.2 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.1 | 0.2 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.1 | 0.6 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.4 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.1 | GO:0042660 | positive regulation of cell fate specification(GO:0042660) |
| 0.0 | 0.1 | GO:1904875 | regulation of DNA ligase activity(GO:1904875) |
| 0.0 | 0.5 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 1.0 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.2 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.2 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 | 0.1 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0090598 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.2 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 | 0.1 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.0 | 0.2 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.2 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.3 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.1 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 | 0.1 | GO:1903898 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) negative regulation of PERK-mediated unfolded protein response(GO:1903898) negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.0 | 0.2 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.3 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.2 | GO:0051459 | regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 | 0.1 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.0 | 0.2 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.6 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.2 | GO:0046449 | creatinine metabolic process(GO:0046449) |
| 0.0 | 0.1 | GO:0045208 | MAPK phosphatase export from nucleus(GO:0045208) MAPK phosphatase export from nucleus, leptomycin B sensitive(GO:0045209) |
| 0.0 | 0.1 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) |
| 0.0 | 0.1 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.0 | 0.3 | GO:0045656 | negative regulation of monocyte differentiation(GO:0045656) |
| 0.0 | 0.1 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.0 | 0.1 | GO:1900148 | Schwann cell proliferation involved in axon regeneration(GO:0014011) negative regulation of Schwann cell migration(GO:1900148) regulation of Schwann cell proliferation involved in axon regeneration(GO:1905044) negative regulation of Schwann cell proliferation involved in axon regeneration(GO:1905045) |
| 0.0 | 0.2 | GO:0071484 | cellular response to light intensity(GO:0071484) |
| 0.0 | 0.1 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.0 | 0.2 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.6 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.2 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.1 | GO:0035766 | cell chemotaxis to fibroblast growth factor(GO:0035766) endothelial cell chemotaxis to fibroblast growth factor(GO:0035768) regulation of cell chemotaxis to fibroblast growth factor(GO:1904847) regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000544) |
| 0.0 | 0.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.1 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.2 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.7 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:0019413 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate metabolic process(GO:0019541) propionate biosynthetic process(GO:0019542) |
| 0.0 | 0.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.1 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.1 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.1 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.0 | 0.0 | GO:0021793 | chemorepulsion of branchiomotor axon(GO:0021793) |
| 0.0 | 0.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.0 | GO:0051102 | DNA ligation involved in DNA recombination(GO:0051102) |
| 0.0 | 0.5 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.1 | GO:0003360 | brainstem development(GO:0003360) |
| 0.0 | 0.1 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.0 | 0.0 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.1 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0070701 | mucus layer(GO:0070701) |
| 0.1 | 0.6 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 0.2 | GO:0031379 | RNA-directed RNA polymerase complex(GO:0031379) |
| 0.1 | 0.6 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.1 | 0.2 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.0 | 0.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.6 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.2 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.4 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.2 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 0.1 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.4 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.1 | GO:0070522 | ERCC4-ERCC1 complex(GO:0070522) |
| 0.0 | 0.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.5 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.5 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 0.2 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.1 | 0.3 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.2 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 0.2 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.0 | 0.2 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.1 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.1 | GO:0097158 | pre-mRNA intronic pyrimidine-rich binding(GO:0097158) |
| 0.0 | 0.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.3 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0004644 | phosphoribosylamine-glycine ligase activity(GO:0004637) phosphoribosylformylglycinamidine cyclo-ligase activity(GO:0004641) phosphoribosylglycinamide formyltransferase activity(GO:0004644) |
| 0.0 | 0.2 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.5 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.1 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 0.6 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.2 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.1 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.0 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.0 | 0.1 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.2 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.3 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.0 | GO:0004423 | iduronate-2-sulfatase activity(GO:0004423) |
| 0.0 | 0.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.1 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.2 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.1 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.2 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.1 | GO:0016933 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.6 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.2 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.5 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.7 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.6 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |