Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
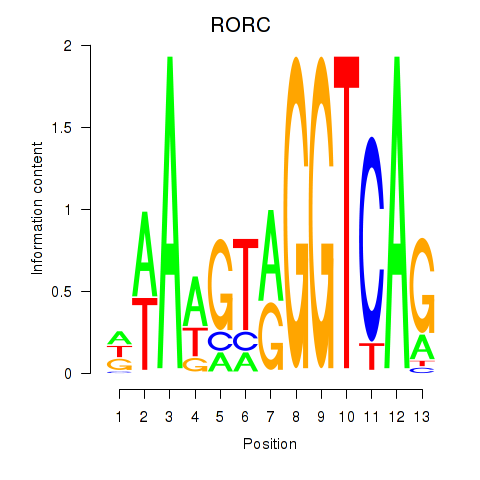
Results for RORC
Z-value: 0.76
Transcription factors associated with RORC
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RORC
|
ENSG00000143365.12 | RAR related orphan receptor C |
Activity profile of RORC motif
Sorted Z-values of RORC motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_64527425 | 0.84 |
ENST00000377432.3
|
PYGM
|
phosphorylase, glycogen, muscle |
| chr19_-_39735646 | 0.82 |
ENST00000413851.2
|
IFNL3
|
interferon, lambda 3 |
| chr19_+_39759154 | 0.80 |
ENST00000331982.5
|
IFNL2
|
interferon, lambda 2 |
| chr8_+_71485681 | 0.55 |
ENST00000391684.1
|
AC120194.1
|
AC120194.1 |
| chr11_-_69867159 | 0.44 |
ENST00000528507.1
|
RP11-626H12.2
|
RP11-626H12.2 |
| chr12_+_93963590 | 0.30 |
ENST00000340600.2
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chr1_-_150207017 | 0.28 |
ENST00000369119.3
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr12_+_93964158 | 0.27 |
ENST00000549206.1
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chr11_+_13299186 | 0.24 |
ENST00000527998.1
ENST00000396441.3 ENST00000533520.1 ENST00000529825.1 ENST00000389707.4 ENST00000401424.1 ENST00000529388.1 ENST00000530357.1 ENST00000403290.1 ENST00000361003.4 ENST00000389708.3 ENST00000403510.3 ENST00000482049.1 |
ARNTL
|
aryl hydrocarbon receptor nuclear translocator-like |
| chr12_+_13061894 | 0.23 |
ENST00000540125.1
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A |
| chrX_+_69501943 | 0.21 |
ENST00000509895.1
ENST00000374473.2 ENST00000276066.4 |
RAB41
|
RAB41, member RAS oncogene family |
| chr14_+_21785693 | 0.21 |
ENST00000382933.4
ENST00000557351.1 |
RPGRIP1
|
retinitis pigmentosa GTPase regulator interacting protein 1 |
| chr1_-_211752073 | 0.20 |
ENST00000367001.4
|
SLC30A1
|
solute carrier family 30 (zinc transporter), member 1 |
| chr4_+_85504075 | 0.19 |
ENST00000295887.5
|
CDS1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1 |
| chr17_-_48785216 | 0.18 |
ENST00000285243.6
|
ANKRD40
|
ankyrin repeat domain 40 |
| chr19_-_7698599 | 0.17 |
ENST00000311069.5
|
PCP2
|
Purkinje cell protein 2 |
| chr19_-_8567505 | 0.15 |
ENST00000600262.1
|
PRAM1
|
PML-RARA regulated adaptor molecule 1 |
| chr3_-_117716418 | 0.15 |
ENST00000484092.1
|
RP11-384F7.2
|
RP11-384F7.2 |
| chr19_-_10450287 | 0.14 |
ENST00000589261.1
ENST00000590569.1 ENST00000589580.1 ENST00000589249.1 |
ICAM3
|
intercellular adhesion molecule 3 |
| chrX_-_43832711 | 0.14 |
ENST00000378062.5
|
NDP
|
Norrie disease (pseudoglioma) |
| chr4_-_170948361 | 0.13 |
ENST00000393702.3
|
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr17_-_42992856 | 0.13 |
ENST00000588316.1
ENST00000435360.2 ENST00000586793.1 ENST00000588735.1 ENST00000588037.1 ENST00000592320.1 ENST00000253408.5 |
GFAP
|
glial fibrillary acidic protein |
| chr12_+_56414795 | 0.13 |
ENST00000431367.2
|
IKZF4
|
IKAROS family zinc finger 4 (Eos) |
| chr14_-_68157084 | 0.13 |
ENST00000557564.1
|
RP11-1012A1.4
|
RP11-1012A1.4 |
| chr1_+_114447763 | 0.13 |
ENST00000369563.3
|
DCLRE1B
|
DNA cross-link repair 1B |
| chr6_+_89855765 | 0.12 |
ENST00000275072.4
|
PM20D2
|
peptidase M20 domain containing 2 |
| chr2_+_162016916 | 0.11 |
ENST00000405852.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr17_-_37764128 | 0.11 |
ENST00000302584.4
|
NEUROD2
|
neuronal differentiation 2 |
| chr6_+_43738444 | 0.11 |
ENST00000324450.6
ENST00000417285.2 ENST00000413642.3 ENST00000372055.4 ENST00000482630.2 ENST00000425836.2 ENST00000372064.4 ENST00000372077.4 ENST00000519767.1 |
VEGFA
|
vascular endothelial growth factor A |
| chr10_-_121296045 | 0.11 |
ENST00000392865.1
|
RGS10
|
regulator of G-protein signaling 10 |
| chr8_+_66955648 | 0.10 |
ENST00000522619.1
|
DNAJC5B
|
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
| chr2_+_162016827 | 0.10 |
ENST00000429217.1
ENST00000406287.1 ENST00000402568.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr7_+_112063192 | 0.10 |
ENST00000005558.4
|
IFRD1
|
interferon-related developmental regulator 1 |
| chr17_+_44679808 | 0.10 |
ENST00000571172.1
|
NSF
|
N-ethylmaleimide-sensitive factor |
| chr2_-_148778323 | 0.09 |
ENST00000440042.1
ENST00000535373.1 ENST00000540442.1 ENST00000536575.1 |
ORC4
|
origin recognition complex, subunit 4 |
| chr19_+_38924316 | 0.09 |
ENST00000355481.4
ENST00000360985.3 ENST00000359596.3 |
RYR1
|
ryanodine receptor 1 (skeletal) |
| chrX_-_49089771 | 0.08 |
ENST00000376251.1
ENST00000323022.5 ENST00000376265.2 |
CACNA1F
|
calcium channel, voltage-dependent, L type, alpha 1F subunit |
| chr3_-_45957534 | 0.08 |
ENST00000536047.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr1_+_236849754 | 0.08 |
ENST00000542672.1
ENST00000366578.4 |
ACTN2
|
actinin, alpha 2 |
| chr2_+_160590469 | 0.08 |
ENST00000409591.1
|
MARCH7
|
membrane-associated ring finger (C3HC4) 7, E3 ubiquitin protein ligase |
| chr22_-_32022280 | 0.08 |
ENST00000442379.1
|
PISD
|
phosphatidylserine decarboxylase |
| chr1_+_159796534 | 0.08 |
ENST00000289707.5
|
SLAMF8
|
SLAM family member 8 |
| chr11_-_10822029 | 0.08 |
ENST00000528839.1
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr3_-_45957088 | 0.08 |
ENST00000539217.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr1_+_26869597 | 0.08 |
ENST00000530003.1
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr8_+_124780672 | 0.07 |
ENST00000521166.1
ENST00000334705.7 |
FAM91A1
|
family with sequence similarity 91, member A1 |
| chr1_-_145826450 | 0.07 |
ENST00000462900.2
|
GPR89A
|
G protein-coupled receptor 89A |
| chr16_+_16326352 | 0.07 |
ENST00000399336.4
ENST00000263012.6 ENST00000538468.1 |
NOMO3
|
NODAL modulator 3 |
| chr2_+_162016804 | 0.06 |
ENST00000392749.2
ENST00000440506.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr17_-_46623441 | 0.05 |
ENST00000330070.4
|
HOXB2
|
homeobox B2 |
| chr21_+_17553910 | 0.05 |
ENST00000428669.2
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr1_-_185286461 | 0.05 |
ENST00000367498.3
|
IVNS1ABP
|
influenza virus NS1A binding protein |
| chr12_+_56624436 | 0.04 |
ENST00000266980.4
ENST00000437277.1 |
SLC39A5
|
solute carrier family 39 (zinc transporter), member 5 |
| chr10_-_103578162 | 0.04 |
ENST00000361464.3
ENST00000357797.5 ENST00000370094.3 |
MGEA5
|
meningioma expressed antigen 5 (hyaluronidase) |
| chr10_-_103578182 | 0.04 |
ENST00000439817.1
|
MGEA5
|
meningioma expressed antigen 5 (hyaluronidase) |
| chr6_-_46922659 | 0.04 |
ENST00000265417.7
|
GPR116
|
G protein-coupled receptor 116 |
| chr3_-_164914640 | 0.04 |
ENST00000241274.3
|
SLITRK3
|
SLIT and NTRK-like family, member 3 |
| chr1_+_154540246 | 0.04 |
ENST00000368476.3
|
CHRNB2
|
cholinergic receptor, nicotinic, beta 2 (neuronal) |
| chr2_-_30144432 | 0.03 |
ENST00000389048.3
|
ALK
|
anaplastic lymphoma receptor tyrosine kinase |
| chr17_-_59940705 | 0.03 |
ENST00000577913.1
|
BRIP1
|
BRCA1 interacting protein C-terminal helicase 1 |
| chr12_+_56623827 | 0.03 |
ENST00000424625.1
ENST00000419753.1 ENST00000454355.2 ENST00000417965.1 ENST00000436633.1 |
SLC39A5
|
solute carrier family 39 (zinc transporter), member 5 |
| chr1_-_217311090 | 0.03 |
ENST00000493603.1
ENST00000366940.2 |
ESRRG
|
estrogen-related receptor gamma |
| chr1_-_62190793 | 0.03 |
ENST00000371177.2
ENST00000606498.1 |
TM2D1
|
TM2 domain containing 1 |
| chr3_-_184095923 | 0.03 |
ENST00000445696.2
ENST00000204615.7 |
THPO
|
thrombopoietin |
| chr10_-_105845674 | 0.02 |
ENST00000353479.5
ENST00000369733.3 |
COL17A1
|
collagen, type XVII, alpha 1 |
| chr4_-_1188922 | 0.02 |
ENST00000515004.1
ENST00000502483.1 |
SPON2
|
spondin 2, extracellular matrix protein |
| chr9_-_110251836 | 0.02 |
ENST00000374672.4
|
KLF4
|
Kruppel-like factor 4 (gut) |
| chr4_+_110749143 | 0.02 |
ENST00000317735.4
|
RRH
|
retinal pigment epithelium-derived rhodopsin homolog |
| chr7_-_43965937 | 0.02 |
ENST00000455877.1
ENST00000223341.7 ENST00000447717.3 ENST00000426198.1 |
URGCP
|
upregulator of cell proliferation |
| chr2_+_148778570 | 0.02 |
ENST00000407073.1
|
MBD5
|
methyl-CpG binding domain protein 5 |
| chr1_+_104198377 | 0.01 |
ENST00000370083.4
|
AMY1A
|
amylase, alpha 1A (salivary) |
| chr18_-_23671139 | 0.01 |
ENST00000579061.1
ENST00000542420.2 |
SS18
|
synovial sarcoma translocation, chromosome 18 |
| chr12_-_39734783 | 0.01 |
ENST00000552961.1
|
KIF21A
|
kinesin family member 21A |
| chr1_+_159796589 | 0.01 |
ENST00000368104.4
|
SLAMF8
|
SLAM family member 8 |
| chr3_+_45986511 | 0.01 |
ENST00000458629.1
ENST00000457814.1 |
CXCR6
|
chemokine (C-X-C motif) receptor 6 |
| chr7_+_20686946 | 0.01 |
ENST00000443026.2
ENST00000406935.1 |
ABCB5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr12_+_7167980 | 0.00 |
ENST00000360817.5
ENST00000402681.3 |
C1S
|
complement component 1, s subcomponent |
Network of associatons between targets according to the STRING database.
First level regulatory network of RORC
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1903568 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.0 | 0.2 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.0 | 0.8 | GO:0002385 | mucosal immune response(GO:0002385) |
| 0.0 | 0.1 | GO:1903572 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.0 | 0.3 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.2 | GO:0097501 | detoxification of inorganic compound(GO:0061687) stress response to metal ion(GO:0097501) |
| 0.0 | 0.6 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.1 | GO:0031860 | telomeric loop formation(GO:0031627) telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.1 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.1 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.0 | 0.8 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.1 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.0 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.0 | 0.1 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990425 | junctional membrane complex(GO:0030314) ryanodine receptor complex(GO:1990425) |
| 0.0 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.3 | GO:0000812 | Swr1 complex(GO:0000812) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.6 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.2 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.3 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.1 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) FATZ binding(GO:0051373) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.2 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.1 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.6 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |