Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
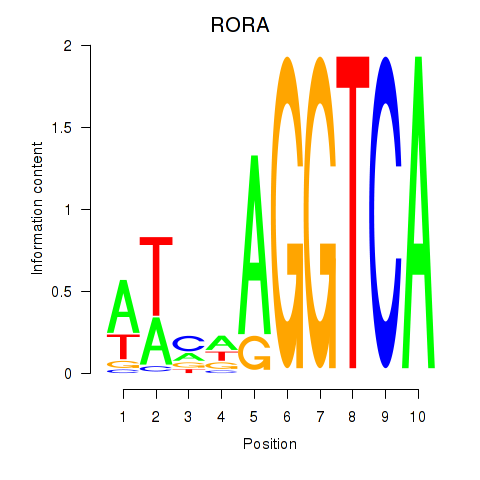
Results for RORA
Z-value: 0.10
Transcription factors associated with RORA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RORA
|
ENSG00000069667.11 | RAR related orphan receptor A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RORA | hg19_v2_chr15_-_60885287_60885325 | -0.82 | 1.8e-01 | Click! |
Activity profile of RORA motif
Sorted Z-values of RORA motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_23791484 | 0.11 |
ENST00000594872.1
|
AL049829.1
|
Uncharacterized protein |
| chr16_+_691792 | 0.08 |
ENST00000307650.4
|
FAM195A
|
family with sequence similarity 195, member A |
| chr5_+_149865377 | 0.07 |
ENST00000522491.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr16_+_1832902 | 0.06 |
ENST00000262302.9
ENST00000563136.1 ENST00000565987.1 ENST00000543305.1 ENST00000568287.1 ENST00000565134.1 |
NUBP2
|
nucleotide binding protein 2 |
| chr16_+_30937213 | 0.06 |
ENST00000427128.1
|
FBXL19
|
F-box and leucine-rich repeat protein 19 |
| chr7_-_150777874 | 0.06 |
ENST00000540185.1
|
FASTK
|
Fas-activated serine/threonine kinase |
| chr11_+_64879317 | 0.06 |
ENST00000526809.1
ENST00000279263.7 ENST00000524986.1 ENST00000534371.1 ENST00000540748.1 ENST00000525385.1 ENST00000345348.5 ENST00000531321.1 ENST00000529414.1 ENST00000526085.1 ENST00000530750.1 |
TM7SF2
|
transmembrane 7 superfamily member 2 |
| chr19_-_49140692 | 0.06 |
ENST00000222122.5
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein |
| chr3_-_149388682 | 0.05 |
ENST00000475579.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr11_+_66624527 | 0.05 |
ENST00000393952.3
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr5_-_126409159 | 0.05 |
ENST00000607731.1
ENST00000535381.1 ENST00000296662.5 ENST00000509733.3 |
C5orf63
|
chromosome 5 open reading frame 63 |
| chrX_-_30327495 | 0.05 |
ENST00000453287.1
|
NR0B1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr8_+_37553261 | 0.05 |
ENST00000331569.4
|
ZNF703
|
zinc finger protein 703 |
| chr16_+_776936 | 0.04 |
ENST00000549114.1
ENST00000341413.4 ENST00000562187.1 ENST00000564537.1 |
HAGHL
|
hydroxyacylglutathione hydrolase-like |
| chr16_-_1821496 | 0.04 |
ENST00000564628.1
ENST00000563498.1 |
NME3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr11_-_118272610 | 0.04 |
ENST00000534438.1
|
RP11-770J1.5
|
Uncharacterized protein |
| chr7_-_128415844 | 0.04 |
ENST00000249389.2
|
OPN1SW
|
opsin 1 (cone pigments), short-wave-sensitive |
| chr10_+_104178946 | 0.04 |
ENST00000432590.1
|
FBXL15
|
F-box and leucine-rich repeat protein 15 |
| chr7_-_150777949 | 0.04 |
ENST00000482571.1
|
FASTK
|
Fas-activated serine/threonine kinase |
| chr19_-_49140609 | 0.04 |
ENST00000601104.1
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein |
| chr19_+_1248547 | 0.04 |
ENST00000586757.1
ENST00000300952.2 |
MIDN
|
midnolin |
| chr17_-_2304365 | 0.04 |
ENST00000575394.1
ENST00000174618.4 |
MNT
|
MAX network transcriptional repressor |
| chr17_+_42148225 | 0.04 |
ENST00000591696.1
|
G6PC3
|
glucose 6 phosphatase, catalytic, 3 |
| chr7_-_150777920 | 0.03 |
ENST00000353841.2
ENST00000297532.6 |
FASTK
|
Fas-activated serine/threonine kinase |
| chr7_-_45151272 | 0.03 |
ENST00000461363.1
ENST00000495078.1 ENST00000494076.1 ENST00000478532.1 ENST00000258770.3 ENST00000361278.3 |
TBRG4
|
transforming growth factor beta regulator 4 |
| chr16_-_88717423 | 0.03 |
ENST00000568278.1
ENST00000569359.1 ENST00000567174.1 |
CYBA
|
cytochrome b-245, alpha polypeptide |
| chr2_-_198364552 | 0.03 |
ENST00000439605.1
ENST00000418022.1 |
HSPD1
|
heat shock 60kDa protein 1 (chaperonin) |
| chr19_-_50311896 | 0.03 |
ENST00000529634.2
|
FUZ
|
fuzzy planar cell polarity protein |
| chr11_-_119599794 | 0.03 |
ENST00000264025.3
|
PVRL1
|
poliovirus receptor-related 1 (herpesvirus entry mediator C) |
| chr19_+_8478154 | 0.03 |
ENST00000381035.4
ENST00000595142.1 ENST00000601724.1 ENST00000393944.1 ENST00000215555.2 ENST00000601283.1 ENST00000595213.1 |
MARCH2
|
membrane-associated ring finger (C3HC4) 2, E3 ubiquitin protein ligase |
| chr5_-_140027175 | 0.03 |
ENST00000512088.1
|
NDUFA2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa |
| chr19_+_54606145 | 0.03 |
ENST00000485876.1
ENST00000391762.1 ENST00000471292.1 ENST00000391763.3 ENST00000391764.3 ENST00000303553.5 |
NDUFA3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3, 9kDa |
| chr16_-_69390209 | 0.03 |
ENST00000563634.1
|
RP11-343C2.9
|
Uncharacterized protein |
| chr3_+_50316458 | 0.03 |
ENST00000316436.3
|
LSMEM2
|
leucine-rich single-pass membrane protein 2 |
| chr22_+_37959647 | 0.03 |
ENST00000415670.1
|
CDC42EP1
|
CDC42 effector protein (Rho GTPase binding) 1 |
| chr12_+_48592134 | 0.03 |
ENST00000595310.1
|
DKFZP779L1853
|
DKFZP779L1853 |
| chr16_+_28303804 | 0.03 |
ENST00000341901.4
|
SBK1
|
SH3 domain binding kinase 1 |
| chr16_+_28875126 | 0.02 |
ENST00000359285.5
ENST00000538342.1 |
SH2B1
|
SH2B adaptor protein 1 |
| chr3_+_46449049 | 0.02 |
ENST00000357392.4
ENST00000400880.3 ENST00000433848.1 |
CCRL2
|
chemokine (C-C motif) receptor-like 2 |
| chr16_+_67563250 | 0.02 |
ENST00000566907.1
|
FAM65A
|
family with sequence similarity 65, member A |
| chr6_-_33297013 | 0.02 |
ENST00000453407.1
|
DAXX
|
death-domain associated protein |
| chr17_-_1928621 | 0.02 |
ENST00000331238.6
|
RTN4RL1
|
reticulon 4 receptor-like 1 |
| chr5_-_140027357 | 0.02 |
ENST00000252102.4
|
NDUFA2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa |
| chr12_+_10366016 | 0.02 |
ENST00000546017.1
ENST00000535576.1 ENST00000539170.1 |
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr17_+_38975358 | 0.02 |
ENST00000436612.1
ENST00000301665.3 |
TMEM99
|
transmembrane protein 99 |
| chr1_-_26197744 | 0.02 |
ENST00000374296.3
|
PAQR7
|
progestin and adipoQ receptor family member VII |
| chr16_+_777739 | 0.02 |
ENST00000563792.1
|
HAGHL
|
hydroxyacylglutathione hydrolase-like |
| chr15_-_72767490 | 0.02 |
ENST00000565181.1
|
RP11-1007O24.3
|
RP11-1007O24.3 |
| chr18_+_9102669 | 0.02 |
ENST00000497577.2
|
NDUFV2
|
NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa |
| chr16_+_29991673 | 0.02 |
ENST00000416441.2
|
TAOK2
|
TAO kinase 2 |
| chr8_-_144655141 | 0.02 |
ENST00000398882.3
|
MROH6
|
maestro heat-like repeat family member 6 |
| chr3_-_50360165 | 0.02 |
ENST00000428028.1
|
HYAL2
|
hyaluronoglucosaminidase 2 |
| chr17_+_48423453 | 0.02 |
ENST00000017003.2
ENST00000509778.1 ENST00000507602.1 |
XYLT2
|
xylosyltransferase II |
| chr1_+_32666188 | 0.02 |
ENST00000421922.2
|
CCDC28B
|
coiled-coil domain containing 28B |
| chr2_+_208414985 | 0.02 |
ENST00000414681.1
|
CREB1
|
cAMP responsive element binding protein 1 |
| chr17_+_74075263 | 0.02 |
ENST00000334586.5
ENST00000392503.2 |
ZACN
|
zinc activated ligand-gated ion channel |
| chr5_+_111964133 | 0.02 |
ENST00000508879.1
ENST00000507565.1 |
RP11-159K7.2
|
RP11-159K7.2 |
| chr16_+_777118 | 0.02 |
ENST00000562141.1
|
HAGHL
|
hydroxyacylglutathione hydrolase-like |
| chr17_-_27503770 | 0.02 |
ENST00000533112.1
|
MYO18A
|
myosin XVIIIA |
| chr1_+_33231268 | 0.02 |
ENST00000373480.1
|
KIAA1522
|
KIAA1522 |
| chr3_-_49158218 | 0.02 |
ENST00000417901.1
ENST00000306026.5 ENST00000434032.2 |
USP19
|
ubiquitin specific peptidase 19 |
| chr11_-_67271723 | 0.02 |
ENST00000533391.1
ENST00000534749.1 ENST00000532703.1 |
PITPNM1
|
phosphatidylinositol transfer protein, membrane-associated 1 |
| chr3_-_123339418 | 0.02 |
ENST00000583087.1
|
MYLK
|
myosin light chain kinase |
| chr16_+_2076869 | 0.01 |
ENST00000424542.2
ENST00000432365.2 |
SLC9A3R2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chr12_-_57039739 | 0.01 |
ENST00000552959.1
ENST00000551020.1 ENST00000553007.2 ENST00000552919.1 ENST00000552104.1 ENST00000262030.3 |
ATP5B
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr9_+_134103496 | 0.01 |
ENST00000498010.1
ENST00000476004.1 ENST00000528406.1 |
NUP214
|
nucleoporin 214kDa |
| chr3_-_49158312 | 0.01 |
ENST00000398892.3
ENST00000453664.1 ENST00000398888.2 |
USP19
|
ubiquitin specific peptidase 19 |
| chr11_+_128563652 | 0.01 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr19_+_7011509 | 0.01 |
ENST00000377296.3
|
AC025278.1
|
Uncharacterized protein |
| chr6_+_36839616 | 0.01 |
ENST00000359359.2
ENST00000510325.2 |
C6orf89
|
chromosome 6 open reading frame 89 |
| chr9_+_74920335 | 0.01 |
ENST00000451596.2
ENST00000436054.1 |
RP11-63P12.6
|
RP11-63P12.6 |
| chr3_-_42845922 | 0.01 |
ENST00000452906.2
|
HIGD1A
|
HIG1 hypoxia inducible domain family, member 1A |
| chr17_-_4458616 | 0.01 |
ENST00000381556.2
|
MYBBP1A
|
MYB binding protein (P160) 1a |
| chr19_-_42931567 | 0.01 |
ENST00000244289.4
|
LIPE
|
lipase, hormone-sensitive |
| chr3_-_52312337 | 0.01 |
ENST00000469000.1
|
WDR82
|
WD repeat domain 82 |
| chr20_-_3748416 | 0.01 |
ENST00000399672.1
|
C20orf27
|
chromosome 20 open reading frame 27 |
| chr19_+_48898132 | 0.01 |
ENST00000263269.3
|
GRIN2D
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2D |
| chr1_+_9294822 | 0.01 |
ENST00000377403.2
|
H6PD
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr10_-_52645416 | 0.01 |
ENST00000374001.2
ENST00000373997.3 ENST00000373995.3 ENST00000282641.2 ENST00000395495.1 ENST00000414883.1 |
A1CF
|
APOBEC1 complementation factor |
| chr20_+_3776371 | 0.01 |
ENST00000245960.5
|
CDC25B
|
cell division cycle 25B |
| chr10_+_99185917 | 0.01 |
ENST00000334828.5
|
PGAM1
|
phosphoglycerate mutase 1 (brain) |
| chr16_-_28303360 | 0.01 |
ENST00000501520.1
|
RP11-57A19.2
|
RP11-57A19.2 |
| chr11_-_129062093 | 0.01 |
ENST00000310343.9
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr19_-_11689752 | 0.01 |
ENST00000592659.1
ENST00000592828.1 ENST00000218758.5 ENST00000412435.2 |
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr12_-_53575120 | 0.01 |
ENST00000542115.1
|
CSAD
|
cysteine sulfinic acid decarboxylase |
| chr6_-_43021437 | 0.01 |
ENST00000265348.3
|
CUL7
|
cullin 7 |
| chr3_-_123339343 | 0.01 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr12_+_72080253 | 0.01 |
ENST00000549735.1
|
TMEM19
|
transmembrane protein 19 |
| chr12_+_65672702 | 0.01 |
ENST00000538045.1
ENST00000535239.1 |
MSRB3
|
methionine sulfoxide reductase B3 |
| chr11_+_86502085 | 0.01 |
ENST00000527521.1
|
PRSS23
|
protease, serine, 23 |
| chr10_+_102729249 | 0.01 |
ENST00000519649.1
ENST00000518124.1 |
SEMA4G
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G |
| chr8_-_144654918 | 0.01 |
ENST00000529971.1
|
MROH6
|
maestro heat-like repeat family member 6 |
| chr10_-_94003003 | 0.01 |
ENST00000412050.4
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr14_-_58893876 | 0.01 |
ENST00000555097.1
ENST00000555404.1 |
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr17_-_2614927 | 0.01 |
ENST00000435359.1
|
CLUH
|
clustered mitochondria (cluA/CLU1) homolog |
| chr12_-_106697974 | 0.01 |
ENST00000553039.1
|
CKAP4
|
cytoskeleton-associated protein 4 |
| chr22_-_38699003 | 0.01 |
ENST00000451964.1
|
CSNK1E
|
casein kinase 1, epsilon |
| chr6_-_107077347 | 0.01 |
ENST00000369063.3
ENST00000539449.1 |
RTN4IP1
|
reticulon 4 interacting protein 1 |
| chr11_+_118272328 | 0.01 |
ENST00000524422.1
|
ATP5L
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G |
| chr19_-_39926268 | 0.01 |
ENST00000599705.1
|
RPS16
|
ribosomal protein S16 |
| chr12_-_16761007 | 0.01 |
ENST00000354662.1
ENST00000441439.2 |
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr2_-_177502659 | 0.01 |
ENST00000295549.4
|
AC017048.3
|
long intergenic non-protein coding RNA 1116 |
| chr17_-_2615031 | 0.01 |
ENST00000576885.1
ENST00000574426.2 |
CLUH
|
clustered mitochondria (cluA/CLU1) homolog |
| chrX_+_10124977 | 0.01 |
ENST00000380833.4
|
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr2_+_219472637 | 0.01 |
ENST00000417849.1
|
PLCD4
|
phospholipase C, delta 4 |
| chr11_+_65292884 | 0.01 |
ENST00000527009.1
|
SCYL1
|
SCY1-like 1 (S. cerevisiae) |
| chr3_+_111753690 | 0.01 |
ENST00000452346.2
|
TMPRSS7
|
transmembrane protease, serine 7 |
| chr19_+_45251804 | 0.01 |
ENST00000164227.5
|
BCL3
|
B-cell CLL/lymphoma 3 |
| chr2_+_134877740 | 0.01 |
ENST00000409645.1
|
MGAT5
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
| chr19_-_42498369 | 0.01 |
ENST00000302102.5
ENST00000545399.1 |
ATP1A3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
| chr3_-_113465065 | 0.01 |
ENST00000497255.1
ENST00000478020.1 ENST00000240922.3 ENST00000493900.1 |
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr16_+_28874860 | 0.01 |
ENST00000545570.1
|
SH2B1
|
SH2B adaptor protein 1 |
| chr16_-_30457048 | 0.01 |
ENST00000500504.2
ENST00000542752.1 |
SEPHS2
|
selenophosphate synthetase 2 |
| chr15_-_53002007 | 0.01 |
ENST00000561490.1
|
FAM214A
|
family with sequence similarity 214, member A |
| chr12_-_58135903 | 0.01 |
ENST00000257897.3
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr3_-_183735651 | 0.01 |
ENST00000427120.2
ENST00000392579.2 ENST00000382494.2 ENST00000446941.2 |
ABCC5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr3_-_24536253 | 0.01 |
ENST00000428492.1
ENST00000396671.2 ENST00000431815.1 ENST00000418247.1 ENST00000416420.1 ENST00000356447.4 |
THRB
|
thyroid hormone receptor, beta |
| chr11_+_86667117 | 0.01 |
ENST00000531827.1
|
RP11-736K20.6
|
RP11-736K20.6 |
| chr12_+_120875887 | 0.01 |
ENST00000229379.2
|
COX6A1
|
cytochrome c oxidase subunit VIa polypeptide 1 |
| chr9_-_39239171 | 0.01 |
ENST00000358144.2
|
CNTNAP3
|
contactin associated protein-like 3 |
| chr6_-_41703296 | 0.01 |
ENST00000373033.1
|
TFEB
|
transcription factor EB |
| chr5_+_32788945 | 0.01 |
ENST00000326958.1
|
AC026703.1
|
AC026703.1 |
| chr12_+_69139886 | 0.00 |
ENST00000398004.2
|
SLC35E3
|
solute carrier family 35, member E3 |
| chr1_+_33231221 | 0.00 |
ENST00000294521.3
|
KIAA1522
|
KIAA1522 |
| chr21_-_43816052 | 0.00 |
ENST00000398405.1
|
TMPRSS3
|
transmembrane protease, serine 3 |
| chr16_-_30537839 | 0.00 |
ENST00000380412.5
|
ZNF768
|
zinc finger protein 768 |
| chr10_+_72575643 | 0.00 |
ENST00000373202.3
|
SGPL1
|
sphingosine-1-phosphate lyase 1 |
| chr17_-_44270133 | 0.00 |
ENST00000262419.6
ENST00000393476.3 |
KANSL1
|
KAT8 regulatory NSL complex subunit 1 |
| chr17_+_2699697 | 0.00 |
ENST00000254695.8
ENST00000366401.4 ENST00000542807.1 |
RAP1GAP2
|
RAP1 GTPase activating protein 2 |
| chr12_-_53320245 | 0.00 |
ENST00000552150.1
|
KRT8
|
keratin 8 |
| chr9_-_35103105 | 0.00 |
ENST00000452248.2
ENST00000356493.5 |
STOML2
|
stomatin (EPB72)-like 2 |
| chr20_-_44485835 | 0.00 |
ENST00000457981.1
ENST00000426915.1 ENST00000217455.4 |
ACOT8
|
acyl-CoA thioesterase 8 |
| chr16_+_31366455 | 0.00 |
ENST00000268296.4
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr7_+_66395596 | 0.00 |
ENST00000424964.2
ENST00000418375.1 |
TMEM248
|
transmembrane protein 248 |
| chr18_-_43678241 | 0.00 |
ENST00000593152.2
ENST00000589252.1 ENST00000590665.1 ENST00000398752.6 |
ATP5A1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle |
| chr1_-_157108266 | 0.00 |
ENST00000326786.4
|
ETV3
|
ets variant 3 |
| chr4_-_186732048 | 0.00 |
ENST00000448662.2
ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr11_+_111957497 | 0.00 |
ENST00000375549.3
ENST00000528182.1 ENST00000528048.1 ENST00000528021.1 ENST00000526592.1 ENST00000525291.1 |
SDHD
|
succinate dehydrogenase complex, subunit D, integral membrane protein |
| chr7_+_129007964 | 0.00 |
ENST00000460109.1
ENST00000474594.1 ENST00000446212.1 |
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr15_+_76352178 | 0.00 |
ENST00000388942.3
|
C15orf27
|
chromosome 15 open reading frame 27 |
| chr17_-_8066250 | 0.00 |
ENST00000488857.1
ENST00000481878.1 ENST00000316509.6 ENST00000498285.1 |
VAMP2
RP11-599B13.6
|
vesicle-associated membrane protein 2 (synaptobrevin 2) Uncharacterized protein |
| chr20_+_37590942 | 0.00 |
ENST00000373325.2
ENST00000252011.3 ENST00000373323.4 |
DHX35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr1_-_153522562 | 0.00 |
ENST00000368714.1
|
S100A4
|
S100 calcium binding protein A4 |
| chr1_+_104293028 | 0.00 |
ENST00000370079.3
|
AMY1C
|
amylase, alpha 1C (salivary) |
| chr9_+_74920408 | 0.00 |
ENST00000451152.1
|
RP11-63P12.6
|
RP11-63P12.6 |
| chr15_-_42565023 | 0.00 |
ENST00000566474.1
|
TMEM87A
|
transmembrane protein 87A |
| chr15_-_75017711 | 0.00 |
ENST00000567032.1
ENST00000564596.1 ENST00000566503.1 ENST00000395049.4 ENST00000395048.2 ENST00000379727.3 |
CYP1A1
|
cytochrome P450, family 1, subfamily A, polypeptide 1 |
| chr17_+_40996590 | 0.00 |
ENST00000253799.3
ENST00000452774.2 |
AOC2
|
amine oxidase, copper containing 2 (retina-specific) |
| chr17_+_4855053 | 0.00 |
ENST00000518175.1
|
ENO3
|
enolase 3 (beta, muscle) |
| chr1_+_206858328 | 0.00 |
ENST00000367103.3
|
MAPKAPK2
|
mitogen-activated protein kinase-activated protein kinase 2 |
| chr3_+_189349162 | 0.00 |
ENST00000264731.3
ENST00000382063.4 ENST00000418709.2 ENST00000320472.5 ENST00000392460.3 ENST00000440651.2 |
TP63
|
tumor protein p63 |
| chr12_+_58013693 | 0.00 |
ENST00000320442.4
ENST00000379218.2 |
SLC26A10
|
solute carrier family 26, member 10 |
| chr12_-_122240792 | 0.00 |
ENST00000545885.1
ENST00000542933.1 ENST00000428029.2 ENST00000541694.1 ENST00000536662.1 ENST00000535643.1 ENST00000541657.1 |
AC084018.1
RHOF
|
AC084018.1 ras homolog family member F (in filopodia) |
| chr9_+_112852477 | 0.00 |
ENST00000480388.1
|
AKAP2
|
A kinase (PRKA) anchor protein 2 |
| chr2_+_171034646 | 0.00 |
ENST00000409044.3
ENST00000408978.4 |
MYO3B
|
myosin IIIB |
| chr6_+_167525277 | 0.00 |
ENST00000400926.2
|
CCR6
|
chemokine (C-C motif) receptor 6 |
| chr16_+_31366536 | 0.00 |
ENST00000562522.1
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr18_-_56296182 | 0.00 |
ENST00000361673.3
|
ALPK2
|
alpha-kinase 2 |
| chr14_-_23755297 | 0.00 |
ENST00000357460.5
|
HOMEZ
|
homeobox and leucine zipper encoding |
| chr2_-_207024134 | 0.00 |
ENST00000457011.1
ENST00000440274.1 ENST00000432169.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr19_-_42498231 | 0.00 |
ENST00000602133.1
|
ATP1A3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
| chr2_+_198365122 | 0.00 |
ENST00000604458.1
|
HSPE1-MOB4
|
HSPE1-MOB4 readthrough |
| chr18_-_12377001 | 0.00 |
ENST00000590811.1
|
AFG3L2
|
AFG3-like AAA ATPase 2 |
| chr12_-_16760021 | 0.00 |
ENST00000540445.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr5_+_132208014 | 0.00 |
ENST00000296877.2
|
LEAP2
|
liver expressed antimicrobial peptide 2 |
| chr3_+_46448648 | 0.00 |
ENST00000399036.3
|
CCRL2
|
chemokine (C-C motif) receptor-like 2 |
| chr17_+_39994032 | 0.00 |
ENST00000293303.4
ENST00000438813.1 |
KLHL10
|
kelch-like family member 10 |
| chr3_-_18480260 | 0.00 |
ENST00000454909.2
|
SATB1
|
SATB homeobox 1 |
| chr8_-_134072593 | 0.00 |
ENST00000427060.2
|
SLA
|
Src-like-adaptor |
| chr5_+_145317356 | 0.00 |
ENST00000511217.1
|
SH3RF2
|
SH3 domain containing ring finger 2 |
| chr15_-_77988485 | 0.00 |
ENST00000561030.1
|
LINGO1
|
leucine rich repeat and Ig domain containing 1 |
| chr10_+_112257596 | 0.00 |
ENST00000369583.3
|
DUSP5
|
dual specificity phosphatase 5 |
| chr11_-_104480019 | 0.00 |
ENST00000536529.1
ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1
|
RP11-886D15.1 |
| chr3_+_128968437 | 0.00 |
ENST00000314797.6
|
COPG1
|
coatomer protein complex, subunit gamma 1 |
| chr18_+_32073253 | 0.00 |
ENST00000283365.9
ENST00000596745.1 ENST00000315456.6 |
DTNA
|
dystrobrevin, alpha |
| chr10_+_77056181 | 0.00 |
ENST00000526759.1
ENST00000533822.1 |
ZNF503-AS1
|
ZNF503 antisense RNA 1 |
| chr16_-_30538079 | 0.00 |
ENST00000562803.1
|
ZNF768
|
zinc finger protein 768 |
| chr16_-_4164027 | 0.00 |
ENST00000572288.1
|
ADCY9
|
adenylate cyclase 9 |
Network of associatons between targets according to the STRING database.
First level regulatory network of RORA
Gene Ontology Analysis
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.0 | 0.1 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.0 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |