Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for RFX5
Z-value: 0.62
Transcription factors associated with RFX5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RFX5
|
ENSG00000143390.13 | regulatory factor X5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RFX5 | hg19_v2_chr1_-_151319710_151319833 | 0.99 | 6.7e-03 | Click! |
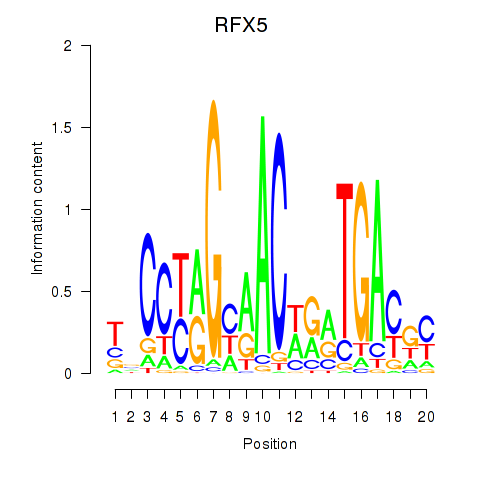
Activity profile of RFX5 motif
Sorted Z-values of RFX5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_51014345 | 0.43 |
ENST00000391815.3
ENST00000594350.1 ENST00000601423.1 |
JOSD2
|
Josephin domain containing 2 |
| chr1_+_249132512 | 0.30 |
ENST00000505503.1
|
ZNF672
|
zinc finger protein 672 |
| chr19_+_50031547 | 0.24 |
ENST00000597801.1
|
RCN3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr1_+_249132462 | 0.21 |
ENST00000306562.3
|
ZNF672
|
zinc finger protein 672 |
| chr6_+_26383318 | 0.21 |
ENST00000469230.1
ENST00000490025.1 ENST00000356709.4 ENST00000352867.2 ENST00000493275.1 ENST00000472507.1 ENST00000482536.1 ENST00000432533.2 ENST00000482842.1 |
BTN2A2
|
butyrophilin, subfamily 2, member A2 |
| chr19_+_36103631 | 0.21 |
ENST00000203166.5
ENST00000379045.2 |
HAUS5
|
HAUS augmin-like complex, subunit 5 |
| chr20_+_306177 | 0.21 |
ENST00000544632.1
|
SOX12
|
SRY (sex determining region Y)-box 12 |
| chr1_-_208417620 | 0.20 |
ENST00000367033.3
|
PLXNA2
|
plexin A2 |
| chr6_+_26440700 | 0.19 |
ENST00000494393.1
ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3
|
butyrophilin, subfamily 3, member A3 |
| chr6_+_32821924 | 0.19 |
ENST00000374859.2
ENST00000453265.2 |
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr6_-_32908792 | 0.16 |
ENST00000418107.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr6_-_32812420 | 0.15 |
ENST00000374881.2
|
PSMB8
|
proteasome (prosome, macropain) subunit, beta type, 8 |
| chr5_-_149792295 | 0.15 |
ENST00000518797.1
ENST00000524315.1 ENST00000009530.7 ENST00000377795.3 |
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain |
| chr20_+_306221 | 0.15 |
ENST00000342665.2
|
SOX12
|
SRY (sex determining region Y)-box 12 |
| chr11_+_66624527 | 0.15 |
ENST00000393952.3
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr6_-_32821599 | 0.14 |
ENST00000354258.4
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr11_-_72433346 | 0.14 |
ENST00000334211.8
|
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr17_-_19265855 | 0.14 |
ENST00000440841.1
ENST00000395615.1 ENST00000461069.2 |
B9D1
|
B9 protein domain 1 |
| chr17_-_19266045 | 0.13 |
ENST00000395616.3
|
B9D1
|
B9 protein domain 1 |
| chr6_+_26383404 | 0.13 |
ENST00000416795.2
ENST00000494184.1 |
BTN2A2
|
butyrophilin, subfamily 2, member A2 |
| chr11_+_45868957 | 0.12 |
ENST00000443527.2
|
CRY2
|
cryptochrome 2 (photolyase-like) |
| chr17_+_38137050 | 0.12 |
ENST00000264639.4
|
PSMD3
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 3 |
| chr12_-_123565834 | 0.11 |
ENST00000546049.1
|
PITPNM2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chr1_+_114473350 | 0.11 |
ENST00000503968.1
|
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr1_-_27339317 | 0.10 |
ENST00000289166.5
|
FAM46B
|
family with sequence similarity 46, member B |
| chr17_-_19265982 | 0.10 |
ENST00000268841.6
ENST00000261499.4 ENST00000575478.1 |
B9D1
|
B9 protein domain 1 |
| chr3_+_9851384 | 0.09 |
ENST00000419081.1
ENST00000438596.1 ENST00000417065.1 ENST00000439814.1 ENST00000418745.1 |
TTLL3
|
tubulin tyrosine ligase-like family, member 3 |
| chr7_+_766320 | 0.08 |
ENST00000297440.6
ENST00000313147.5 |
HEATR2
|
HEAT repeat containing 2 |
| chr6_-_30181133 | 0.08 |
ENST00000454678.2
ENST00000434785.1 |
TRIM26
|
tripartite motif containing 26 |
| chr6_+_26365387 | 0.07 |
ENST00000532865.1
ENST00000530653.1 ENST00000527417.1 |
BTN3A2
|
butyrophilin, subfamily 3, member A2 |
| chr6_-_30658745 | 0.06 |
ENST00000376420.5
ENST00000376421.5 |
NRM
|
nurim (nuclear envelope membrane protein) |
| chr6_-_32731299 | 0.06 |
ENST00000435145.2
ENST00000437316.2 |
HLA-DQB2
|
major histocompatibility complex, class II, DQ beta 2 |
| chr6_-_30181156 | 0.06 |
ENST00000418026.1
ENST00000416596.1 ENST00000453195.1 |
TRIM26
|
tripartite motif containing 26 |
| chr11_-_118436707 | 0.06 |
ENST00000264020.2
ENST00000264021.3 |
IFT46
|
intraflagellar transport 46 homolog (Chlamydomonas) |
| chr16_-_74808710 | 0.06 |
ENST00000219368.3
ENST00000544337.1 |
FA2H
|
fatty acid 2-hydroxylase |
| chr8_-_99306611 | 0.06 |
ENST00000341166.3
|
NIPAL2
|
NIPA-like domain containing 2 |
| chr19_+_19496624 | 0.06 |
ENST00000494516.2
ENST00000360315.3 ENST00000252577.5 |
GATAD2A
|
GATA zinc finger domain containing 2A |
| chr4_-_83719884 | 0.05 |
ENST00000282709.4
ENST00000273908.4 |
SCD5
|
stearoyl-CoA desaturase 5 |
| chr5_-_55412774 | 0.05 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chr19_+_45417921 | 0.05 |
ENST00000252491.4
ENST00000592885.1 ENST00000589781.1 |
APOC1
|
apolipoprotein C-I |
| chr3_+_9850199 | 0.05 |
ENST00000452597.1
|
TTLL3
|
tubulin tyrosine ligase-like family, member 3 |
| chr1_+_181003067 | 0.05 |
ENST00000434571.2
ENST00000367579.3 ENST00000282990.6 ENST00000367580.5 |
MR1
|
major histocompatibility complex, class I-related |
| chr13_-_30424821 | 0.05 |
ENST00000380680.4
|
UBL3
|
ubiquitin-like 3 |
| chr3_+_66271489 | 0.05 |
ENST00000536651.1
|
SLC25A26
|
solute carrier family 25 (S-adenosylmethionine carrier), member 26 |
| chr19_+_45418067 | 0.04 |
ENST00000589078.1
ENST00000586638.1 |
APOC1
|
apolipoprotein C-I |
| chr10_-_82116505 | 0.04 |
ENST00000372202.1
ENST00000421924.2 ENST00000453477.1 |
DYDC1
|
DPY30 domain containing 1 |
| chr6_-_32920794 | 0.04 |
ENST00000395305.3
ENST00000395303.3 ENST00000374843.4 ENST00000429234.1 |
HLA-DMA
XXbac-BPG181M17.5
|
major histocompatibility complex, class II, DM alpha Uncharacterized protein |
| chr8_-_99306564 | 0.04 |
ENST00000430223.2
|
NIPAL2
|
NIPA-like domain containing 2 |
| chr3_+_66271410 | 0.04 |
ENST00000336733.6
|
SLC25A26
|
solute carrier family 25 (S-adenosylmethionine carrier), member 26 |
| chr1_-_202936394 | 0.04 |
ENST00000367249.4
|
CYB5R1
|
cytochrome b5 reductase 1 |
| chr8_-_98290087 | 0.04 |
ENST00000322128.3
|
TSPYL5
|
TSPY-like 5 |
| chr4_-_143226979 | 0.03 |
ENST00000514525.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr1_+_85527987 | 0.03 |
ENST00000326813.8
ENST00000294664.6 ENST00000528899.1 |
WDR63
|
WD repeat domain 63 |
| chr22_-_30987837 | 0.03 |
ENST00000335214.6
|
PES1
|
pescadillo ribosomal biogenesis factor 1 |
| chr10_-_14880002 | 0.03 |
ENST00000465530.1
|
CDNF
|
cerebral dopamine neurotrophic factor |
| chr12_-_110841462 | 0.03 |
ENST00000455511.3
ENST00000450008.2 |
ANAPC7
|
anaphase promoting complex subunit 7 |
| chr11_+_8008867 | 0.02 |
ENST00000309828.4
ENST00000449102.2 |
EIF3F
|
eukaryotic translation initiation factor 3, subunit F |
| chr6_+_26365443 | 0.02 |
ENST00000527422.1
ENST00000356386.2 ENST00000396934.3 ENST00000377708.2 ENST00000396948.1 ENST00000508906.2 |
BTN3A2
|
butyrophilin, subfamily 3, member A2 |
| chr2_-_242088850 | 0.02 |
ENST00000358649.4
ENST00000452907.1 ENST00000403638.3 ENST00000539818.1 ENST00000415234.1 ENST00000234040.4 |
PASK
|
PAS domain containing serine/threonine kinase |
| chr3_-_169587621 | 0.01 |
ENST00000523069.1
ENST00000316428.5 ENST00000264676.5 |
LRRC31
|
leucine rich repeat containing 31 |
| chr6_+_26458171 | 0.01 |
ENST00000493173.1
ENST00000541522.1 ENST00000429381.1 ENST00000469185.1 |
BTN2A1
|
butyrophilin, subfamily 2, member A1 |
| chr17_+_38137073 | 0.01 |
ENST00000541736.1
|
PSMD3
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 3 |
| chr19_+_10527449 | 0.00 |
ENST00000592685.1
ENST00000380702.2 |
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chr6_+_32812568 | 0.00 |
ENST00000414474.1
|
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr19_+_19496728 | 0.00 |
ENST00000537887.1
ENST00000417582.2 |
GATAD2A
|
GATA zinc finger domain containing 2A |
Network of associatons between targets according to the STRING database.
First level regulatory network of RFX5
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0002503 | MHC class II protein complex assembly(GO:0002399) peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.0 | 0.1 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.1 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.0 | 0.2 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.1 | GO:0010900 | negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 0.0 | 0.1 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.0 | 0.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.2 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.0 | 0.1 | GO:0070736 | protein-glycine ligase activity, initiating(GO:0070736) |
| 0.0 | 0.1 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.1 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.1 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 0.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |