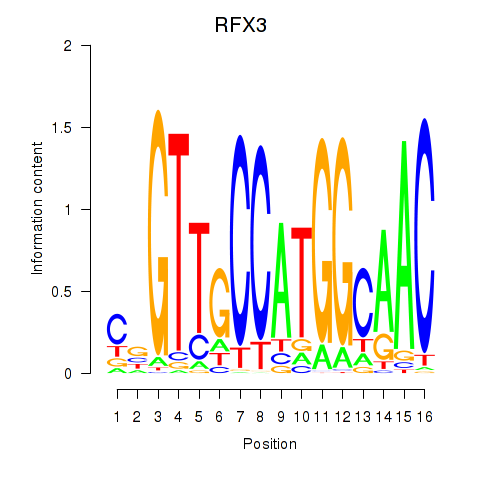
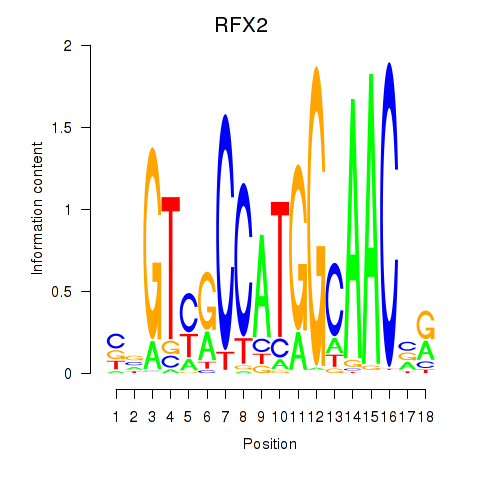
|
chrX_-_48693955
Show fit
|
1.46 |
ENST00000218230.5
|
PCSK1N
|
proprotein convertase subtilisin/kexin type 1 inhibitor
|
|
chr2_-_74648702
Show fit
|
1.42 |
ENST00000518863.1
|
C2orf81
|
chromosome 2 open reading frame 81
|
|
chr19_-_59084647
Show fit
|
1.31 |
ENST00000594234.1
ENST00000596039.1
|
MZF1
|
myeloid zinc finger 1
|
|
chr12_-_64062583
Show fit
|
0.72 |
ENST00000542209.1
|
DPY19L2
|
dpy-19-like 2 (C. elegans)
|
|
chr19_-_51017881
Show fit
|
0.71 |
ENST00000601207.1
ENST00000598657.1
ENST00000376916.3
|
ASPDH
|
aspartate dehydrogenase domain containing
|
|
chr4_+_147096837
Show fit
|
0.68 |
ENST00000296581.5
ENST00000502781.1
|
LSM6
|
LSM6 homolog, U6 small nuclear RNA associated (S. cerevisiae)
|
|
chr15_-_52971544
Show fit
|
0.67 |
ENST00000566768.1
ENST00000561543.1
|
FAM214A
|
family with sequence similarity 214, member A
|
|
chr19_-_58892389
Show fit
|
0.65 |
ENST00000427624.2
ENST00000597582.1
|
ZNF837
|
zinc finger protein 837
|
|
chr16_-_30773372
Show fit
|
0.65 |
ENST00000545825.1
ENST00000541260.1
|
C16orf93
|
chromosome 16 open reading frame 93
|
|
chr19_+_1261106
Show fit
|
0.64 |
ENST00000588411.1
|
CIRBP
|
cold inducible RNA binding protein
|
|
chr19_-_59084922
Show fit
|
0.63 |
ENST00000215057.2
ENST00000599369.1
|
MZF1
|
myeloid zinc finger 1
|
|
chr19_+_17413663
Show fit
|
0.59 |
ENST00000594999.1
|
MRPL34
|
mitochondrial ribosomal protein L34
|
|
chr10_+_104178946
Show fit
|
0.57 |
ENST00000432590.1
|
FBXL15
|
F-box and leucine-rich repeat protein 15
|
|
chr19_-_7553889
Show fit
|
0.55 |
ENST00000221480.1
|
PEX11G
|
peroxisomal biogenesis factor 11 gamma
|
|
chr11_-_64885111
Show fit
|
0.54 |
ENST00000528598.1
ENST00000310597.4
|
ZNHIT2
|
zinc finger, HIT-type containing 2
|
|
chr19_-_17356697
Show fit
|
0.54 |
ENST00000291442.3
|
NR2F6
|
nuclear receptor subfamily 2, group F, member 6
|
|
chr22_-_38349552
Show fit
|
0.53 |
ENST00000422191.1
ENST00000249079.2
ENST00000418863.1
ENST00000403305.1
ENST00000403026.1
|
C22orf23
|
chromosome 22 open reading frame 23
|
|
chr3_-_42003613
Show fit
|
0.51 |
ENST00000414606.1
|
ULK4
|
unc-51 like kinase 4
|
|
chr1_+_161068179
Show fit
|
0.48 |
ENST00000368011.4
ENST00000392192.2
|
KLHDC9
|
kelch domain containing 9
|
|
chr19_+_5681153
Show fit
|
0.48 |
ENST00000579559.1
ENST00000577222.1
|
HSD11B1L
RPL36
|
hydroxysteroid (11-beta) dehydrogenase 1-like
ribosomal protein L36
|
|
chr9_+_131085095
Show fit
|
0.47 |
ENST00000372875.3
|
COQ4
|
coenzyme Q4
|
|
chr19_-_7553852
Show fit
|
0.47 |
ENST00000593547.1
|
PEX11G
|
peroxisomal biogenesis factor 11 gamma
|
|
chr17_+_38375528
Show fit
|
0.46 |
ENST00000583268.1
|
WIPF2
|
WAS/WASL interacting protein family, member 2
|
|
chr19_-_55791058
Show fit
|
0.45 |
ENST00000587959.1
ENST00000585927.1
ENST00000587922.1
ENST00000585698.1
|
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1
|
|
chr5_+_61708488
Show fit
|
0.45 |
ENST00000505902.1
|
IPO11
|
importin 11
|
|
chr3_-_118753792
Show fit
|
0.45 |
ENST00000480431.1
|
IGSF11
|
immunoglobulin superfamily, member 11
|
|
chr14_+_78227105
Show fit
|
0.44 |
ENST00000439131.2
ENST00000355883.3
ENST00000557011.1
ENST00000556047.1
|
C14orf178
|
chromosome 14 open reading frame 178
|
|
chr2_+_74682150
Show fit
|
0.43 |
ENST00000233331.7
ENST00000431187.1
ENST00000409917.1
ENST00000409493.2
|
INO80B
|
INO80 complex subunit B
|
|
chr11_-_57089774
Show fit
|
0.43 |
ENST00000527207.1
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa
|
|
chr19_+_18530184
Show fit
|
0.42 |
ENST00000601357.2
|
SSBP4
|
single stranded DNA binding protein 4
|
|
chr6_-_43484621
Show fit
|
0.41 |
ENST00000506469.1
ENST00000503972.1
|
YIPF3
|
Yip1 domain family, member 3
|
|
chr19_+_58962971
Show fit
|
0.41 |
ENST00000336614.4
ENST00000545523.1
ENST00000599194.1
ENST00000598244.1
ENST00000599193.1
ENST00000594214.1
ENST00000391696.1
|
ZNF324B
|
zinc finger protein 324B
|
|
chr10_-_103347883
Show fit
|
0.41 |
ENST00000339310.3
ENST00000370158.3
ENST00000299206.4
ENST00000456836.2
ENST00000413344.1
ENST00000429502.1
ENST00000430045.1
ENST00000370172.1
ENST00000436284.2
ENST00000370162.3
|
POLL
|
polymerase (DNA directed), lambda
|
|
chr11_+_66360665
Show fit
|
0.40 |
ENST00000310190.4
|
CCS
|
copper chaperone for superoxide dismutase
|
|
chr9_+_136325089
Show fit
|
0.40 |
ENST00000291722.7
ENST00000316948.4
ENST00000540581.1
|
CACFD1
|
calcium channel flower domain containing 1
|
|
chr17_-_8079632
Show fit
|
0.40 |
ENST00000431792.2
|
TMEM107
|
transmembrane protein 107
|
|
chr22_-_38240316
Show fit
|
0.40 |
ENST00000411961.2
ENST00000434930.1
|
ANKRD54
|
ankyrin repeat domain 54
|
|
chr11_+_537494
Show fit
|
0.40 |
ENST00000270115.7
|
LRRC56
|
leucine rich repeat containing 56
|
|
chr11_+_844406
Show fit
|
0.40 |
ENST00000397404.1
|
TSPAN4
|
tetraspanin 4
|
|
chr12_+_48577366
Show fit
|
0.40 |
ENST00000316554.3
|
C12orf68
|
chromosome 12 open reading frame 68
|
|
chr19_+_50529329
Show fit
|
0.39 |
ENST00000599155.1
|
ZNF473
|
zinc finger protein 473
|
|
chr1_-_16563641
Show fit
|
0.38 |
ENST00000375599.3
|
RSG1
|
REM2 and RAB-like small GTPase 1
|
|
chr16_+_57662527
Show fit
|
0.38 |
ENST00000563374.1
ENST00000568234.1
ENST00000565770.1
ENST00000564338.1
ENST00000566164.1
|
GPR56
|
G protein-coupled receptor 56
|
|
chr9_+_35829208
Show fit
|
0.37 |
ENST00000439587.2
ENST00000377991.4
|
TMEM8B
|
transmembrane protein 8B
|
|
chr12_-_110486281
Show fit
|
0.36 |
ENST00000546627.1
|
C12orf76
|
chromosome 12 open reading frame 76
|
|
chr5_-_54529415
Show fit
|
0.36 |
ENST00000282572.4
|
CCNO
|
cyclin O
|
|
chr19_+_1450112
Show fit
|
0.36 |
ENST00000590469.1
ENST00000233607.2
ENST00000238483.4
ENST00000590877.1
|
APC2
|
adenomatosis polyposis coli 2
|
|
chr2_+_220462560
Show fit
|
0.35 |
ENST00000456909.1
ENST00000295641.10
|
STK11IP
|
serine/threonine kinase 11 interacting protein
|
|
chr16_+_30671223
Show fit
|
0.35 |
ENST00000568722.1
|
FBRS
|
fibrosin
|
|
chr17_-_7761256
Show fit
|
0.35 |
ENST00000575208.1
|
LSMD1
|
LSM domain containing 1
|
|
chr19_-_41870026
Show fit
|
0.35 |
ENST00000243578.3
|
B9D2
|
B9 protein domain 2
|
|
chr16_+_57662596
Show fit
|
0.34 |
ENST00000567397.1
ENST00000568979.1
|
GPR56
|
G protein-coupled receptor 56
|
|
chr4_+_141445333
Show fit
|
0.34 |
ENST00000507667.1
|
ELMOD2
|
ELMO/CED-12 domain containing 2
|
|
chr1_-_156571254
Show fit
|
0.34 |
ENST00000438976.2
ENST00000334588.7
ENST00000368232.4
ENST00000415314.2
|
GPATCH4
|
G patch domain containing 4
|
|
chr16_-_67450325
Show fit
|
0.34 |
ENST00000348579.2
|
ZDHHC1
|
zinc finger, DHHC-type containing 1
|
|
chr22_-_38240412
Show fit
|
0.33 |
ENST00000215941.4
|
ANKRD54
|
ankyrin repeat domain 54
|
|
chr19_-_40324767
Show fit
|
0.33 |
ENST00000601972.1
ENST00000430012.2
ENST00000323039.5
ENST00000348817.3
|
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B
|
|
chr12_+_4671352
Show fit
|
0.33 |
ENST00000542744.1
|
DYRK4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4
|
|
chr20_+_43992094
Show fit
|
0.33 |
ENST00000453003.1
|
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae)
|
|
chr5_+_176873789
Show fit
|
0.33 |
ENST00000323249.3
ENST00000502922.1
|
PRR7
|
proline rich 7 (synaptic)
|
|
chr11_-_27384737
Show fit
|
0.32 |
ENST00000317945.6
|
CCDC34
|
coiled-coil domain containing 34
|
|
chr1_+_179051160
Show fit
|
0.32 |
ENST00000367625.4
ENST00000352445.6
|
TOR3A
|
torsin family 3, member A
|
|
chr2_+_219221573
Show fit
|
0.32 |
ENST00000289388.3
|
C2orf62
|
chromosome 2 open reading frame 62
|
|
chr11_-_75379479
Show fit
|
0.32 |
ENST00000434603.2
|
MAP6
|
microtubule-associated protein 6
|
|
chr17_-_61523622
Show fit
|
0.31 |
ENST00000448884.2
ENST00000582297.1
ENST00000582034.1
ENST00000578072.1
ENST00000360793.3
|
CYB561
|
cytochrome b561
|
|
chrX_+_102024075
Show fit
|
0.31 |
ENST00000431616.1
ENST00000440496.1
ENST00000420471.1
ENST00000435966.1
|
LINC00630
|
long intergenic non-protein coding RNA 630
|
|
chr1_-_167905225
Show fit
|
0.31 |
ENST00000367846.4
|
MPC2
|
mitochondrial pyruvate carrier 2
|
|
chr22_+_30752606
Show fit
|
0.31 |
ENST00000399824.2
ENST00000405659.1
ENST00000338306.3
|
CCDC157
|
coiled-coil domain containing 157
|
|
chr11_-_8615720
Show fit
|
0.30 |
ENST00000358872.3
ENST00000454443.2
|
STK33
|
serine/threonine kinase 33
|
|
chr2_+_159651821
Show fit
|
0.30 |
ENST00000309950.3
ENST00000409042.1
|
DAPL1
|
death associated protein-like 1
|
|
chr12_+_6930964
Show fit
|
0.30 |
ENST00000382315.3
|
GPR162
|
G protein-coupled receptor 162
|
|
chr3_+_187420101
Show fit
|
0.30 |
ENST00000449623.1
ENST00000437407.1
|
RP11-211G3.3
|
Uncharacterized protein
|
|
chr16_+_30773610
Show fit
|
0.30 |
ENST00000566811.1
|
RNF40
|
ring finger protein 40, E3 ubiquitin protein ligase
|
|
chr9_-_130497565
Show fit
|
0.30 |
ENST00000336067.6
ENST00000373281.5
ENST00000373284.5
ENST00000458505.3
|
TOR2A
|
torsin family 2, member A
|
|
chr14_+_105992906
Show fit
|
0.30 |
ENST00000392519.2
|
TMEM121
|
transmembrane protein 121
|
|
chr19_+_50879705
Show fit
|
0.29 |
ENST00000598168.1
ENST00000411902.2
ENST00000253727.5
ENST00000597790.1
ENST00000597130.1
ENST00000599105.1
|
NR1H2
|
nuclear receptor subfamily 1, group H, member 2
|
|
chr21_-_43916433
Show fit
|
0.29 |
ENST00000291536.3
|
RSPH1
|
radial spoke head 1 homolog (Chlamydomonas)
|
|
chr19_+_18530146
Show fit
|
0.29 |
ENST00000348495.6
ENST00000270061.7
|
SSBP4
|
single stranded DNA binding protein 4
|
|
chr6_-_43484718
Show fit
|
0.28 |
ENST00000372422.2
|
YIPF3
|
Yip1 domain family, member 3
|
|
chr7_-_156433195
Show fit
|
0.28 |
ENST00000333319.6
|
C7orf13
|
chromosome 7 open reading frame 13
|
|
chr12_-_2966193
Show fit
|
0.28 |
ENST00000382678.3
|
AC005841.1
|
Uncharacterized protein ENSP00000372125
|
|
chr1_+_43637996
Show fit
|
0.28 |
ENST00000528956.1
ENST00000529956.1
|
WDR65
|
WD repeat domain 65
|
|
chr19_+_9945962
Show fit
|
0.28 |
ENST00000587625.1
ENST00000247970.4
ENST00000588695.1
|
PIN1
|
peptidylprolyl cis/trans isomerase, NIMA-interacting 1
|
|
chr17_-_41322332
Show fit
|
0.28 |
ENST00000590740.1
|
RP11-242D8.1
|
RP11-242D8.1
|
|
chr6_+_30848740
Show fit
|
0.28 |
ENST00000505534.1
|
DDR1
|
discoidin domain receptor tyrosine kinase 1
|
|
chr6_+_31939608
Show fit
|
0.27 |
ENST00000375331.2
ENST00000375333.2
|
STK19
|
serine/threonine kinase 19
|
|
chr17_+_1944790
Show fit
|
0.27 |
ENST00000575162.1
|
DPH1
|
diphthamide biosynthesis 1
|
|
chr22_-_31063782
Show fit
|
0.26 |
ENST00000404885.1
ENST00000403268.1
ENST00000407308.1
ENST00000342474.4
ENST00000334679.3
|
DUSP18
|
dual specificity phosphatase 18
|
|
chrX_+_153146127
Show fit
|
0.26 |
ENST00000452593.1
ENST00000357566.1
|
LCA10
|
Putative lung carcinoma-associated protein 10
|
|
chr19_+_19639670
Show fit
|
0.26 |
ENST00000436027.5
|
YJEFN3
|
YjeF N-terminal domain containing 3
|
|
chr19_-_6393216
Show fit
|
0.26 |
ENST00000595047.1
|
GTF2F1
|
general transcription factor IIF, polypeptide 1, 74kDa
|
|
chr1_+_161129240
Show fit
|
0.26 |
ENST00000492950.1
|
USP21
|
ubiquitin specific peptidase 21
|
|
chr8_+_144798429
Show fit
|
0.26 |
ENST00000338033.4
ENST00000395107.4
ENST00000395108.2
|
MAPK15
|
mitogen-activated protein kinase 15
|
|
chr19_-_46974741
Show fit
|
0.26 |
ENST00000313683.10
ENST00000602246.1
|
PNMAL1
|
paraneoplastic Ma antigen family-like 1
|
|
chr7_+_73275483
Show fit
|
0.26 |
ENST00000320531.2
|
WBSCR28
|
Williams-Beuren syndrome chromosome region 28
|
|
chr7_-_1177874
Show fit
|
0.26 |
ENST00000397098.3
ENST00000357429.6
ENST00000397100.2
ENST00000491163.1
|
C7orf50
|
chromosome 7 open reading frame 50
|
|
chr11_+_61447845
Show fit
|
0.25 |
ENST00000257215.5
|
DAGLA
|
diacylglycerol lipase, alpha
|
|
chr9_-_131418944
Show fit
|
0.25 |
ENST00000419989.1
ENST00000451652.1
ENST00000372715.2
|
WDR34
|
WD repeat domain 34
|
|
chr17_-_7760779
Show fit
|
0.25 |
ENST00000335155.5
ENST00000575071.1
|
LSMD1
|
LSM domain containing 1
|
|
chr19_-_7990991
Show fit
|
0.24 |
ENST00000318978.4
|
CTXN1
|
cortexin 1
|
|
chr17_+_6544078
Show fit
|
0.24 |
ENST00000250101.5
|
TXNDC17
|
thioredoxin domain containing 17
|
|
chr6_+_30848771
Show fit
|
0.24 |
ENST00000503180.1
|
DDR1
|
discoidin domain receptor tyrosine kinase 1
|
|
chr7_+_102073966
Show fit
|
0.24 |
ENST00000495936.1
ENST00000356387.2
ENST00000478730.2
ENST00000468241.1
ENST00000403646.3
|
ORAI2
|
ORAI calcium release-activated calcium modulator 2
|
|
chr19_+_56652556
Show fit
|
0.24 |
ENST00000337080.3
|
ZNF444
|
zinc finger protein 444
|
|
chr7_+_150725510
Show fit
|
0.24 |
ENST00000461373.1
ENST00000358849.4
ENST00000297504.6
ENST00000542328.1
ENST00000498578.1
ENST00000356058.4
ENST00000477719.1
ENST00000477092.1
|
ABCB8
|
ATP-binding cassette, sub-family B (MDR/TAP), member 8
|
|
chr14_-_74181106
Show fit
|
0.24 |
ENST00000316836.3
|
PNMA1
|
paraneoplastic Ma antigen 1
|
|
chr3_+_149192475
Show fit
|
0.24 |
ENST00000465758.1
|
TM4SF4
|
transmembrane 4 L six family member 4
|
|
chr7_+_150065278
Show fit
|
0.24 |
ENST00000519397.1
ENST00000479668.1
ENST00000540729.1
|
REPIN1
|
replication initiator 1
|
|
chr17_-_73840614
Show fit
|
0.24 |
ENST00000586108.1
|
UNC13D
|
unc-13 homolog D (C. elegans)
|
|
chr19_-_5680499
Show fit
|
0.23 |
ENST00000587589.1
|
C19orf70
|
chromosome 19 open reading frame 70
|
|
chr19_-_5680891
Show fit
|
0.23 |
ENST00000309324.4
|
C19orf70
|
chromosome 19 open reading frame 70
|
|
chr8_-_101964738
Show fit
|
0.23 |
ENST00000523938.1
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta
|
|
chr1_-_21978312
Show fit
|
0.23 |
ENST00000359708.4
ENST00000290101.4
|
RAP1GAP
|
RAP1 GTPase activating protein
|
|
chr2_-_203735586
Show fit
|
0.23 |
ENST00000454326.1
ENST00000432273.1
ENST00000450143.1
ENST00000411681.1
|
ICA1L
|
islet cell autoantigen 1,69kDa-like
|
|
chr11_-_67271723
Show fit
|
0.23 |
ENST00000533391.1
ENST00000534749.1
ENST00000532703.1
|
PITPNM1
|
phosphatidylinositol transfer protein, membrane-associated 1
|
|
chr19_-_1490398
Show fit
|
0.23 |
ENST00000588671.1
ENST00000300954.5
|
PCSK4
|
proprotein convertase subtilisin/kexin type 4
|
|
chr19_+_56165480
Show fit
|
0.23 |
ENST00000450554.2
|
U2AF2
|
U2 small nuclear RNA auxiliary factor 2
|
|
chr7_+_150065879
Show fit
|
0.23 |
ENST00000397281.2
ENST00000444957.1
ENST00000466559.1
ENST00000489432.2
ENST00000475514.1
ENST00000482680.1
ENST00000488943.1
ENST00000518514.1
ENST00000478789.1
|
REPIN1
ZNF775
|
replication initiator 1
zinc finger protein 775
|
|
chr6_+_144980954
Show fit
|
0.23 |
ENST00000367525.3
|
UTRN
|
utrophin
|
|
chr7_+_100797678
Show fit
|
0.23 |
ENST00000337619.5
|
AP1S1
|
adaptor-related protein complex 1, sigma 1 subunit
|
|
chr14_+_24025345
Show fit
|
0.23 |
ENST00000557630.1
|
THTPA
|
thiamine triphosphatase
|
|
chr1_+_45274154
Show fit
|
0.22 |
ENST00000450269.1
ENST00000453418.1
ENST00000409335.2
|
BTBD19
|
BTB (POZ) domain containing 19
|
|
chr19_-_633576
Show fit
|
0.22 |
ENST00000588649.2
|
POLRMT
|
polymerase (RNA) mitochondrial (DNA directed)
|
|
chr7_+_99699179
Show fit
|
0.22 |
ENST00000438383.1
ENST00000429084.1
ENST00000359593.4
ENST00000439416.1
|
AP4M1
|
adaptor-related protein complex 4, mu 1 subunit
|
|
chr6_+_30848557
Show fit
|
0.22 |
ENST00000460944.2
ENST00000324771.8
|
DDR1
|
discoidin domain receptor tyrosine kinase 1
|
|
chr17_+_6544356
Show fit
|
0.22 |
ENST00000574838.1
|
TXNDC17
|
thioredoxin domain containing 17
|
|
chr16_-_30064244
Show fit
|
0.22 |
ENST00000571269.1
ENST00000561666.1
|
FAM57B
|
family with sequence similarity 57, member B
|
|
chr6_+_30848829
Show fit
|
0.22 |
ENST00000508317.1
|
DDR1
|
discoidin domain receptor tyrosine kinase 1
|
|
chr19_+_55851221
Show fit
|
0.22 |
ENST00000255613.3
ENST00000539076.1
|
SUV420H2
AC020922.1
|
suppressor of variegation 4-20 homolog 2 (Drosophila)
Uncharacterized protein
|
|
chr11_-_66104237
Show fit
|
0.22 |
ENST00000530056.1
|
RIN1
|
Ras and Rab interactor 1
|
|
chr19_+_56652643
Show fit
|
0.22 |
ENST00000586123.1
|
ZNF444
|
zinc finger protein 444
|
|
chr17_+_26662679
Show fit
|
0.21 |
ENST00000578158.1
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial)
|
|
chr1_-_156217829
Show fit
|
0.21 |
ENST00000356983.2
ENST00000335852.1
ENST00000340183.5
ENST00000540423.1
|
PAQR6
|
progestin and adipoQ receptor family member VI
|
|
chr1_-_36916066
Show fit
|
0.21 |
ENST00000315643.9
|
OSCP1
|
organic solute carrier partner 1
|
|
chr1_+_155178481
Show fit
|
0.21 |
ENST00000368376.3
|
MTX1
|
metaxin 1
|
|
chr3_-_129147432
Show fit
|
0.21 |
ENST00000503957.1
ENST00000505956.1
ENST00000326085.3
|
EFCAB12
|
EF-hand calcium binding domain 12
|
|
chr16_-_54963026
Show fit
|
0.21 |
ENST00000560208.1
ENST00000557792.1
|
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding)
|
|
chr19_+_14063278
Show fit
|
0.21 |
ENST00000254337.6
|
DCAF15
|
DDB1 and CUL4 associated factor 15
|
|
chr19_-_46405861
Show fit
|
0.21 |
ENST00000322217.5
|
MYPOP
|
Myb-related transcription factor, partner of profilin
|
|
chrX_+_103173457
Show fit
|
0.20 |
ENST00000419165.1
|
TMSB15B
|
thymosin beta 15B
|
|
chr22_-_50746027
Show fit
|
0.20 |
ENST00000425954.1
ENST00000449103.1
|
PLXNB2
|
plexin B2
|
|
chr3_-_10052849
Show fit
|
0.20 |
ENST00000437616.1
ENST00000429065.2
|
AC022007.5
|
AC022007.5
|
|
chr9_+_131084846
Show fit
|
0.20 |
ENST00000608951.1
|
COQ4
|
coenzyme Q4
|
|
chr19_+_19639704
Show fit
|
0.20 |
ENST00000514277.4
|
YJEFN3
|
YjeF N-terminal domain containing 3
|
|
chr1_+_44457261
Show fit
|
0.20 |
ENST00000372318.3
|
CCDC24
|
coiled-coil domain containing 24
|
|
chr16_+_28986134
Show fit
|
0.20 |
ENST00000352260.7
|
SPNS1
|
spinster homolog 1 (Drosophila)
|
|
chr20_+_42984330
Show fit
|
0.20 |
ENST00000316673.4
ENST00000609795.1
ENST00000457232.1
ENST00000609262.1
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr17_-_7761172
Show fit
|
0.20 |
ENST00000333775.5
ENST00000575771.1
|
LSMD1
|
LSM domain containing 1
|
|
chr1_+_74663994
Show fit
|
0.20 |
ENST00000472069.1
|
FPGT
|
fucose-1-phosphate guanylyltransferase
|
|
chr11_+_66278080
Show fit
|
0.20 |
ENST00000318312.7
ENST00000526815.1
ENST00000537537.1
ENST00000525809.1
ENST00000455748.2
ENST00000393994.2
|
BBS1
|
Bardet-Biedl syndrome 1
|
|
chr5_+_121647877
Show fit
|
0.20 |
ENST00000514497.2
ENST00000261367.7
|
SNCAIP
|
synuclein, alpha interacting protein
|
|
chr3_-_107941209
Show fit
|
0.20 |
ENST00000492106.1
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas)
|
|
chr19_-_42759300
Show fit
|
0.20 |
ENST00000222329.4
|
ERF
|
Ets2 repressor factor
|
|
chr2_-_130902567
Show fit
|
0.20 |
ENST00000457413.1
ENST00000392984.3
ENST00000409128.1
ENST00000441670.1
ENST00000409943.3
ENST00000409234.3
ENST00000310463.6
|
CCDC74B
|
coiled-coil domain containing 74B
|
|
chr21_-_44846999
Show fit
|
0.20 |
ENST00000270162.6
|
SIK1
|
salt-inducible kinase 1
|
|
chr16_+_30772913
Show fit
|
0.19 |
ENST00000563909.1
|
RNF40
|
ring finger protein 40, E3 ubiquitin protein ligase
|
|
chr10_-_135150367
Show fit
|
0.19 |
ENST00000368555.3
ENST00000252939.4
ENST00000368558.1
ENST00000368556.2
|
CALY
|
calcyon neuron-specific vesicular protein
|
|
chr7_+_102389434
Show fit
|
0.19 |
ENST00000409231.3
ENST00000418198.1
|
FAM185A
|
family with sequence similarity 185, member A
|
|
chr11_-_57089671
Show fit
|
0.19 |
ENST00000532437.1
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa
|
|
chr1_+_183605200
Show fit
|
0.19 |
ENST00000304685.4
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr19_-_58874112
Show fit
|
0.19 |
ENST00000311044.3
ENST00000595763.1
ENST00000425453.3
|
ZNF497
|
zinc finger protein 497
|
|
chr3_-_49761337
Show fit
|
0.19 |
ENST00000535833.1
ENST00000308388.6
ENST00000480687.1
ENST00000308375.6
|
AMIGO3
GMPPB
|
adhesion molecule with Ig-like domain 3
GDP-mannose pyrophosphorylase B
|
|
chr12_+_56511943
Show fit
|
0.19 |
ENST00000257940.2
ENST00000552345.1
ENST00000551880.1
ENST00000546903.1
ENST00000551790.1
|
ZC3H10
ESYT1
|
zinc finger CCCH-type containing 10
extended synaptotagmin-like protein 1
|
|
chr14_+_105941118
Show fit
|
0.19 |
ENST00000550577.1
ENST00000538259.2
|
CRIP2
|
cysteine-rich protein 2
|
|
chr19_+_54371114
Show fit
|
0.19 |
ENST00000448420.1
ENST00000439000.1
ENST00000391770.4
ENST00000391771.1
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr11_-_61129723
Show fit
|
0.19 |
ENST00000537680.1
ENST00000426130.2
ENST00000294072.4
|
CYB561A3
|
cytochrome b561 family, member A3
|
|
chr19_-_40950182
Show fit
|
0.19 |
ENST00000596456.1
|
SERTAD3
|
SERTA domain containing 3
|
|
chr6_+_35227449
Show fit
|
0.19 |
ENST00000373953.3
ENST00000440666.2
ENST00000339411.5
|
ZNF76
|
zinc finger protein 76
|
|
chr22_+_30752963
Show fit
|
0.18 |
ENST00000445005.1
ENST00000430839.1
|
CCDC157
|
coiled-coil domain containing 157
|
|
chr15_-_68497657
Show fit
|
0.18 |
ENST00000448060.2
ENST00000467889.1
|
CALML4
|
calmodulin-like 4
|
|
chr19_-_58919815
Show fit
|
0.18 |
ENST00000597980.1
|
CTD-2619J13.14
|
CTD-2619J13.14
|
|
chr19_-_47249679
Show fit
|
0.18 |
ENST00000263280.6
|
STRN4
|
striatin, calmodulin binding protein 4
|
|
chr12_+_7014126
Show fit
|
0.18 |
ENST00000415834.1
ENST00000436789.1
|
LRRC23
|
leucine rich repeat containing 23
|
|
chr11_-_559377
Show fit
|
0.18 |
ENST00000486629.1
|
C11orf35
|
chromosome 11 open reading frame 35
|
|
chr18_+_77905794
Show fit
|
0.18 |
ENST00000587254.1
ENST00000586421.1
|
AC139100.2
|
Uncharacterized protein
|
|
chr2_+_27193480
Show fit
|
0.18 |
ENST00000233121.2
ENST00000405074.3
|
MAPRE3
|
microtubule-associated protein, RP/EB family, member 3
|
|
chr8_+_38243821
Show fit
|
0.18 |
ENST00000519476.2
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2
|
|
chr2_+_220144168
Show fit
|
0.18 |
ENST00000392087.2
ENST00000442681.1
ENST00000439026.1
|
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2
|
|
chr16_-_57836321
Show fit
|
0.18 |
ENST00000569112.1
ENST00000562311.1
ENST00000445690.2
ENST00000379655.4
|
KIFC3
|
kinesin family member C3
|
|
chr19_-_50370799
Show fit
|
0.18 |
ENST00000600910.1
ENST00000322344.3
ENST00000600573.1
|
PNKP
|
polynucleotide kinase 3'-phosphatase
|
|
chr9_-_131085021
Show fit
|
0.18 |
ENST00000372890.4
|
TRUB2
|
TruB pseudouridine (psi) synthase family member 2
|
|
chr16_-_71264558
Show fit
|
0.17 |
ENST00000448089.2
ENST00000393550.2
ENST00000448691.1
ENST00000393567.2
ENST00000321489.5
ENST00000539973.1
ENST00000288168.10
ENST00000545267.1
ENST00000541601.1
ENST00000538248.1
|
HYDIN
|
HYDIN, axonemal central pair apparatus protein
|
|
chr17_+_4736627
Show fit
|
0.17 |
ENST00000355280.6
ENST00000347992.7
|
MINK1
|
misshapen-like kinase 1
|
|
chr6_-_31940065
Show fit
|
0.17 |
ENST00000375349.3
ENST00000337523.5
|
DXO
|
decapping exoribonuclease
|
|
chr22_+_41601209
Show fit
|
0.17 |
ENST00000216237.5
|
L3MBTL2
|
l(3)mbt-like 2 (Drosophila)
|
|
chr22_-_33968239
Show fit
|
0.17 |
ENST00000452586.2
ENST00000421768.1
|
LARGE
|
like-glycosyltransferase
|
|
chr17_+_78389247
Show fit
|
0.17 |
ENST00000520136.2
ENST00000520284.1
ENST00000517795.1
ENST00000523228.1
ENST00000523828.1
ENST00000522200.1
ENST00000521565.1
ENST00000518907.1
ENST00000518644.1
ENST00000518901.1
|
ENDOV
|
endonuclease V
|
|
chr19_+_35630628
Show fit
|
0.17 |
ENST00000588715.1
ENST00000588607.1
|
FXYD1
|
FXYD domain containing ion transport regulator 1
|
|
chr5_-_142065612
Show fit
|
0.17 |
ENST00000360966.5
ENST00000411960.1
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr21_+_18885318
Show fit
|
0.17 |
ENST00000400166.1
|
CXADR
|
coxsackie virus and adenovirus receptor
|
|
chr16_-_88923285
Show fit
|
0.17 |
ENST00000542788.1
ENST00000569433.1
ENST00000268695.5
ENST00000568311.1
|
GALNS
|
galactosamine (N-acetyl)-6-sulfate sulfatase
|
|
chr15_+_42066888
Show fit
|
0.17 |
ENST00000510535.1
|
MAPKBP1
|
mitogen-activated protein kinase binding protein 1
|
|
chr8_-_135725205
Show fit
|
0.17 |
ENST00000523399.1
ENST00000377838.3
|
ZFAT
|
zinc finger and AT hook domain containing
|
|
chr19_-_51289436
Show fit
|
0.17 |
ENST00000562076.1
|
CTD-2568A17.1
|
CTD-2568A17.1
|
|
chr12_-_110486348
Show fit
|
0.17 |
ENST00000547573.1
ENST00000546651.2
ENST00000551185.2
|
C12orf76
|
chromosome 12 open reading frame 76
|
|
chr18_+_44526786
Show fit
|
0.16 |
ENST00000245121.5
ENST00000356157.7
|
KATNAL2
|
katanin p60 subunit A-like 2
|
|
chr7_+_99699280
Show fit
|
0.16 |
ENST00000421755.1
|
AP4M1
|
adaptor-related protein complex 4, mu 1 subunit
|
|
chr2_-_203736452
Show fit
|
0.16 |
ENST00000419460.1
|
ICA1L
|
islet cell autoantigen 1,69kDa-like
|
|
chr17_-_8079648
Show fit
|
0.16 |
ENST00000449985.2
ENST00000532998.1
ENST00000437139.2
ENST00000533070.1
ENST00000316425.5
|
TMEM107
|
transmembrane protein 107
|
|
chr15_-_55790515
Show fit
|
0.16 |
ENST00000448430.2
ENST00000457155.2
|
DYX1C1
|
dyslexia susceptibility 1 candidate 1
|