Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
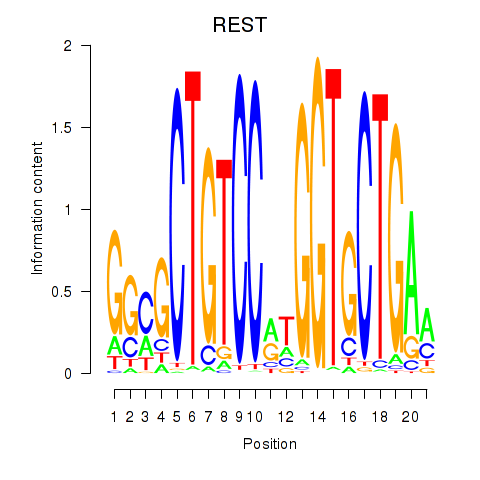
Results for REST
Z-value: 0.76
Transcription factors associated with REST
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
REST
|
ENSG00000084093.11 | RE1 silencing transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| REST | hg19_v2_chr4_+_57774042_57774114 | -0.17 | 8.3e-01 | Click! |
Activity profile of REST motif
Sorted Z-values of REST motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_31199037 | 0.61 |
ENST00000424224.1
|
OSBP2
|
oxysterol binding protein 2 |
| chr10_+_99609996 | 0.44 |
ENST00000370602.1
|
GOLGA7B
|
golgin A7 family, member B |
| chr14_+_96039882 | 0.43 |
ENST00000556346.1
ENST00000553785.1 |
RP11-1070N10.4
|
RP11-1070N10.4 |
| chr12_+_52695617 | 0.40 |
ENST00000293525.5
|
KRT86
|
keratin 86 |
| chr19_+_39759154 | 0.38 |
ENST00000331982.5
|
IFNL2
|
interferon, lambda 2 |
| chr19_-_40724246 | 0.36 |
ENST00000311308.6
|
TTC9B
|
tetratricopeptide repeat domain 9B |
| chr12_-_12837423 | 0.36 |
ENST00000540510.1
|
GPR19
|
G protein-coupled receptor 19 |
| chr20_+_62327996 | 0.33 |
ENST00000369996.1
|
TNFRSF6B
|
tumor necrosis factor receptor superfamily, member 6b, decoy |
| chr2_-_27531313 | 0.27 |
ENST00000296099.2
|
UCN
|
urocortin |
| chrX_-_153141434 | 0.27 |
ENST00000407935.2
ENST00000439496.1 |
L1CAM
|
L1 cell adhesion molecule |
| chr19_-_55691614 | 0.24 |
ENST00000592470.1
ENST00000354308.3 |
SYT5
|
synaptotagmin V |
| chr17_+_30813576 | 0.23 |
ENST00000313401.3
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
| chr14_-_94759595 | 0.21 |
ENST00000261994.4
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr3_+_193853927 | 0.20 |
ENST00000232424.3
|
HES1
|
hes family bHLH transcription factor 1 |
| chr14_-_94759361 | 0.20 |
ENST00000393096.1
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr14_-_94759408 | 0.19 |
ENST00000554723.1
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr1_+_33352036 | 0.19 |
ENST00000373467.3
|
HPCA
|
hippocalcin |
| chr19_-_55691472 | 0.19 |
ENST00000537500.1
|
SYT5
|
synaptotagmin V |
| chr1_+_154540246 | 0.18 |
ENST00000368476.3
|
CHRNB2
|
cholinergic receptor, nicotinic, beta 2 (neuronal) |
| chr19_-_55691377 | 0.18 |
ENST00000589172.1
|
SYT5
|
synaptotagmin V |
| chr15_+_75287861 | 0.17 |
ENST00000425597.3
ENST00000562327.1 ENST00000568018.1 ENST00000562212.1 ENST00000567920.1 ENST00000566872.1 ENST00000361900.6 ENST00000545456.1 |
SCAMP5
|
secretory carrier membrane protein 5 |
| chr2_-_51259641 | 0.17 |
ENST00000406316.2
ENST00000405581.1 |
NRXN1
|
neurexin 1 |
| chr2_+_226265364 | 0.16 |
ENST00000272907.6
|
NYAP2
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2 |
| chr11_-_75141206 | 0.16 |
ENST00000376292.4
|
KLHL35
|
kelch-like family member 35 |
| chrX_+_153237740 | 0.15 |
ENST00000369982.4
|
TMEM187
|
transmembrane protein 187 |
| chrX_-_47479246 | 0.14 |
ENST00000295987.7
ENST00000340666.4 |
SYN1
|
synapsin I |
| chr11_+_64808675 | 0.14 |
ENST00000529996.1
|
SAC3D1
|
SAC3 domain containing 1 |
| chr10_-_102790852 | 0.13 |
ENST00000470414.1
ENST00000370215.3 |
PDZD7
|
PDZ domain containing 7 |
| chr12_+_119419294 | 0.12 |
ENST00000267260.4
|
SRRM4
|
serine/arginine repetitive matrix 4 |
| chr12_-_99288536 | 0.12 |
ENST00000549797.1
ENST00000333732.7 ENST00000341752.7 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr3_+_9745510 | 0.12 |
ENST00000383831.3
|
CPNE9
|
copine family member IX |
| chr6_+_43603552 | 0.12 |
ENST00000372171.4
|
MAD2L1BP
|
MAD2L1 binding protein |
| chr1_+_212606219 | 0.11 |
ENST00000366988.3
|
NENF
|
neudesin neurotrophic factor |
| chr19_-_11639910 | 0.11 |
ENST00000588998.1
ENST00000586149.1 |
ECSIT
|
ECSIT signalling integrator |
| chr11_-_66115032 | 0.11 |
ENST00000311181.4
|
B3GNT1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1 |
| chrX_+_153238220 | 0.10 |
ENST00000425274.1
|
TMEM187
|
transmembrane protein 187 |
| chr12_+_71833550 | 0.10 |
ENST00000266674.5
|
LGR5
|
leucine-rich repeat containing G protein-coupled receptor 5 |
| chr3_+_9745487 | 0.10 |
ENST00000383832.3
|
CPNE9
|
copine family member IX |
| chr19_-_45927097 | 0.10 |
ENST00000340192.7
|
ERCC1
|
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) |
| chr3_+_62304712 | 0.09 |
ENST00000494481.1
|
C3orf14
|
chromosome 3 open reading frame 14 |
| chr19_-_45927622 | 0.09 |
ENST00000300853.3
ENST00000589165.1 |
ERCC1
|
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) |
| chr12_+_113796347 | 0.09 |
ENST00000545182.2
ENST00000280800.3 |
PLBD2
|
phospholipase B domain containing 2 |
| chr15_-_72612470 | 0.08 |
ENST00000287202.5
|
CELF6
|
CUGBP, Elav-like family member 6 |
| chr8_-_145752390 | 0.08 |
ENST00000529415.2
ENST00000533758.1 |
LRRC24
|
leucine rich repeat containing 24 |
| chr6_+_154360616 | 0.08 |
ENST00000229768.5
ENST00000419506.2 ENST00000524163.1 ENST00000414028.2 ENST00000435918.2 ENST00000337049.4 |
OPRM1
|
opioid receptor, mu 1 |
| chr9_-_139096955 | 0.08 |
ENST00000371748.5
|
LHX3
|
LIM homeobox 3 |
| chr19_-_11639931 | 0.08 |
ENST00000592312.1
ENST00000590480.1 ENST00000585318.1 ENST00000252440.7 ENST00000417981.2 ENST00000270517.7 |
ECSIT
|
ECSIT signalling integrator |
| chr13_-_111214015 | 0.07 |
ENST00000267328.3
|
RAB20
|
RAB20, member RAS oncogene family |
| chr5_-_176037105 | 0.07 |
ENST00000303991.4
|
GPRIN1
|
G protein regulated inducer of neurite outgrowth 1 |
| chr16_+_3019246 | 0.07 |
ENST00000318782.8
ENST00000293978.8 |
PAQR4
|
progestin and adipoQ receptor family member IV |
| chr18_+_57567180 | 0.06 |
ENST00000316660.6
ENST00000269518.9 |
PMAIP1
|
phorbol-12-myristate-13-acetate-induced protein 1 |
| chr10_+_102790980 | 0.06 |
ENST00000393459.1
ENST00000224807.5 |
SFXN3
|
sideroflexin 3 |
| chrX_-_153141302 | 0.06 |
ENST00000361699.4
ENST00000543994.1 ENST00000370057.3 ENST00000538883.1 ENST00000361981.3 |
L1CAM
|
L1 cell adhesion molecule |
| chr19_-_51845378 | 0.06 |
ENST00000335624.4
|
VSIG10L
|
V-set and immunoglobulin domain containing 10 like |
| chr19_-_45926739 | 0.06 |
ENST00000589381.1
ENST00000591636.1 ENST00000013807.5 ENST00000592023.1 |
ERCC1
|
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) |
| chr16_+_5008290 | 0.05 |
ENST00000251170.7
|
SEC14L5
|
SEC14-like 5 (S. cerevisiae) |
| chr20_+_30555805 | 0.05 |
ENST00000562532.2
|
XKR7
|
XK, Kell blood group complex subunit-related family, member 7 |
| chr2_-_47798044 | 0.05 |
ENST00000327876.4
|
KCNK12
|
potassium channel, subfamily K, member 12 |
| chr1_-_40105617 | 0.04 |
ENST00000372852.3
|
HEYL
|
hes-related family bHLH transcription factor with YRPW motif-like |
| chr20_-_62284766 | 0.04 |
ENST00000370053.1
|
STMN3
|
stathmin-like 3 |
| chr1_+_171750776 | 0.03 |
ENST00000458517.1
ENST00000362019.3 ENST00000367737.5 ENST00000361735.3 |
METTL13
|
methyltransferase like 13 |
| chrX_-_153237258 | 0.03 |
ENST00000310441.7
|
HCFC1
|
host cell factor C1 (VP16-accessory protein) |
| chr10_+_89419370 | 0.03 |
ENST00000361175.4
ENST00000456849.1 |
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr17_-_27332931 | 0.03 |
ENST00000442608.3
ENST00000335960.6 |
SEZ6
|
seizure related 6 homolog (mouse) |
| chr7_-_65113280 | 0.03 |
ENST00000593865.1
|
AC104057.1
|
Protein LOC100996407 |
| chr6_+_158957431 | 0.03 |
ENST00000367090.3
|
TMEM181
|
transmembrane protein 181 |
| chr11_+_45907177 | 0.03 |
ENST00000241014.2
|
MAPK8IP1
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chr10_-_50970382 | 0.03 |
ENST00000419399.1
ENST00000432695.1 |
OGDHL
|
oxoglutarate dehydrogenase-like |
| chr10_-_50970322 | 0.02 |
ENST00000374103.4
|
OGDHL
|
oxoglutarate dehydrogenase-like |
| chr2_+_46524537 | 0.02 |
ENST00000263734.3
|
EPAS1
|
endothelial PAS domain protein 1 |
| chr17_-_27333163 | 0.02 |
ENST00000360295.9
|
SEZ6
|
seizure related 6 homolog (mouse) |
| chr17_-_27333311 | 0.02 |
ENST00000317338.12
ENST00000585644.1 |
SEZ6
|
seizure related 6 homolog (mouse) |
| chr2_+_115199876 | 0.02 |
ENST00000436732.1
ENST00000410059.1 |
DPP10
|
dipeptidyl-peptidase 10 (non-functional) |
| chr15_+_68117872 | 0.02 |
ENST00000380035.2
ENST00000389002.1 |
SKOR1
|
SKI family transcriptional corepressor 1 |
| chr20_+_44657845 | 0.02 |
ENST00000243964.3
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5 |
| chr19_-_3971050 | 0.01 |
ENST00000545797.2
ENST00000596311.1 |
DAPK3
|
death-associated protein kinase 3 |
| chr14_-_51561784 | 0.01 |
ENST00000360392.4
|
TRIM9
|
tripartite motif containing 9 |
| chr3_+_62304648 | 0.01 |
ENST00000462069.1
ENST00000232519.5 ENST00000465142.1 |
C3orf14
|
chromosome 3 open reading frame 14 |
| chr2_-_51259528 | 0.01 |
ENST00000404971.1
|
NRXN1
|
neurexin 1 |
| chr11_-_92931098 | 0.01 |
ENST00000326402.4
|
SLC36A4
|
solute carrier family 36 (proton/amino acid symporter), member 4 |
| chr12_-_99288986 | 0.01 |
ENST00000552407.1
ENST00000551613.1 ENST00000548447.1 ENST00000546364.3 ENST00000552748.1 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr10_+_60936921 | 0.01 |
ENST00000373878.3
|
PHYHIPL
|
phytanoyl-CoA 2-hydroxylase interacting protein-like |
| chr12_-_57443886 | 0.01 |
ENST00000300119.3
|
MYO1A
|
myosin IA |
| chr8_-_91095099 | 0.01 |
ENST00000265431.3
|
CALB1
|
calbindin 1, 28kDa |
| chr6_+_25652432 | 0.00 |
ENST00000377961.2
|
SCGN
|
secretagogin, EF-hand calcium binding protein |
| chr15_+_68117844 | 0.00 |
ENST00000554054.1
|
SKOR1
|
SKI family transcriptional corepressor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of REST
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.1 | 0.6 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.2 | GO:0021722 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.1 | 0.2 | GO:0060164 | negative regulation of auditory receptor cell differentiation(GO:0045608) negative regulation of mechanoreceptor differentiation(GO:0045632) regulation of timing of neuron differentiation(GO:0060164) negative regulation of inner ear receptor cell differentiation(GO:2000981) |
| 0.0 | 0.2 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.2 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.4 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.2 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.0 | 0.2 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.0 | 0.1 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.1 | GO:0032097 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 0.0 | 0.1 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.4 | GO:0002385 | mucosal immune response(GO:0002385) |
| 0.0 | 0.0 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 0.0 | 0.1 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.0 | 0.1 | GO:0032099 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.1 | 0.2 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.1 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.0 | 0.2 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.4 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.7 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.2 | GO:0044327 | dendritic spine head(GO:0044327) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.1 | 0.2 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.0 | 0.2 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.4 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.0 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.0 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |