Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
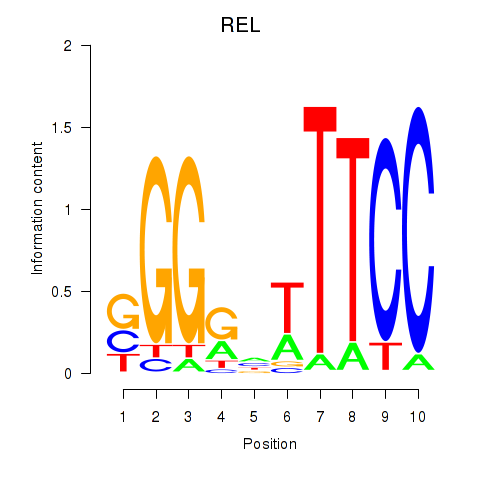
Results for REL
Z-value: 0.45
Transcription factors associated with REL
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
REL
|
ENSG00000162924.9 | REL proto-oncogene, NF-kB subunit |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| REL | hg19_v2_chr2_+_61108650_61108687 | -1.00 | 4.4e-03 | Click! |
Activity profile of REL motif
Sorted Z-values of REL motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_46272106 | 0.42 |
ENST00000560168.1
|
SIX5
|
SIX homeobox 5 |
| chr19_-_46272462 | 0.40 |
ENST00000317578.6
|
SIX5
|
SIX homeobox 5 |
| chr12_+_51632638 | 0.37 |
ENST00000549732.2
|
DAZAP2
|
DAZ associated protein 2 |
| chrX_-_48755030 | 0.29 |
ENST00000490755.2
ENST00000465150.2 ENST00000495490.2 |
TIMM17B
|
translocase of inner mitochondrial membrane 17 homolog B (yeast) |
| chr19_-_47734448 | 0.29 |
ENST00000439096.2
|
BBC3
|
BCL2 binding component 3 |
| chr16_-_88717482 | 0.26 |
ENST00000261623.3
|
CYBA
|
cytochrome b-245, alpha polypeptide |
| chr22_-_39268192 | 0.25 |
ENST00000216083.6
|
CBX6
|
chromobox homolog 6 |
| chr14_-_69262947 | 0.24 |
ENST00000557086.1
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr11_-_66313699 | 0.24 |
ENST00000526986.1
ENST00000310442.3 |
ZDHHC24
|
zinc finger, DHHC-type containing 24 |
| chr1_+_201979743 | 0.23 |
ENST00000446188.1
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr12_+_51632666 | 0.22 |
ENST00000604900.1
|
DAZAP2
|
DAZ associated protein 2 |
| chr2_+_219264466 | 0.22 |
ENST00000273062.2
|
CTDSP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 |
| chr11_-_65381643 | 0.21 |
ENST00000309100.3
ENST00000529839.1 ENST00000526293.1 |
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11 |
| chr20_-_36156125 | 0.21 |
ENST00000397135.1
ENST00000397137.1 |
BLCAP
|
bladder cancer associated protein |
| chr9_-_34637806 | 0.20 |
ENST00000477726.1
|
SIGMAR1
|
sigma non-opioid intracellular receptor 1 |
| chr14_-_69262916 | 0.20 |
ENST00000553375.1
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr19_-_46145696 | 0.20 |
ENST00000588172.1
|
EML2
|
echinoderm microtubule associated protein like 2 |
| chr3_+_183967409 | 0.20 |
ENST00000324557.4
ENST00000402825.3 |
ECE2
|
endothelin converting enzyme 2 |
| chr11_+_119039414 | 0.19 |
ENST00000409991.1
ENST00000292199.2 ENST00000409265.4 ENST00000409109.1 |
NLRX1
|
NLR family member X1 |
| chr22_-_39268308 | 0.19 |
ENST00000407418.3
|
CBX6
|
chromobox homolog 6 |
| chr17_-_73511584 | 0.19 |
ENST00000321617.3
|
CASKIN2
|
CASK interacting protein 2 |
| chr2_-_128145498 | 0.18 |
ENST00000409179.2
|
MAP3K2
|
mitogen-activated protein kinase kinase kinase 2 |
| chr16_+_2820912 | 0.18 |
ENST00000570539.1
|
SRRM2
|
serine/arginine repetitive matrix 2 |
| chr14_+_103589789 | 0.17 |
ENST00000558056.1
ENST00000560869.1 |
TNFAIP2
|
tumor necrosis factor, alpha-induced protein 2 |
| chr9_-_136344197 | 0.17 |
ENST00000414172.1
ENST00000371897.4 |
SLC2A6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr2_+_220094657 | 0.17 |
ENST00000436226.1
|
ANKZF1
|
ankyrin repeat and zinc finger domain containing 1 |
| chr21_+_46875424 | 0.16 |
ENST00000359759.4
|
COL18A1
|
collagen, type XVIII, alpha 1 |
| chr19_-_6591113 | 0.16 |
ENST00000423145.3
ENST00000245903.3 |
CD70
|
CD70 molecule |
| chr12_-_62997214 | 0.15 |
ENST00000408887.2
|
C12orf61
|
chromosome 12 open reading frame 61 |
| chr6_-_32122106 | 0.15 |
ENST00000428778.1
|
PRRT1
|
proline-rich transmembrane protein 1 |
| chr12_+_51633061 | 0.15 |
ENST00000551313.1
|
DAZAP2
|
DAZ associated protein 2 |
| chr20_-_36156264 | 0.15 |
ENST00000445723.1
ENST00000414080.1 |
BLCAP
|
bladder cancer associated protein |
| chr17_-_4852332 | 0.14 |
ENST00000572383.1
|
PFN1
|
profilin 1 |
| chr3_-_156878482 | 0.14 |
ENST00000295925.4
|
CCNL1
|
cyclin L1 |
| chr1_+_110453203 | 0.13 |
ENST00000357302.4
ENST00000344188.5 ENST00000329608.6 |
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr11_+_119038897 | 0.13 |
ENST00000454811.1
ENST00000449394.1 |
NLRX1
|
NLR family member X1 |
| chr20_-_36156293 | 0.13 |
ENST00000373537.2
ENST00000414542.2 |
BLCAP
|
bladder cancer associated protein |
| chr17_+_46189311 | 0.13 |
ENST00000582481.1
|
SNX11
|
sorting nexin 11 |
| chr2_+_87808725 | 0.13 |
ENST00000413202.1
|
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr1_+_45274154 | 0.13 |
ENST00000450269.1
ENST00000453418.1 ENST00000409335.2 |
BTBD19
|
BTB (POZ) domain containing 19 |
| chr19_+_11466062 | 0.13 |
ENST00000251473.5
ENST00000591329.1 ENST00000586380.1 |
DKFZP761J1410
|
Lipid phosphate phosphatase-related protein type 2 |
| chr12_+_51632600 | 0.12 |
ENST00000549555.1
ENST00000439799.2 ENST00000425012.2 |
DAZAP2
|
DAZ associated protein 2 |
| chr17_+_25799008 | 0.12 |
ENST00000583370.1
ENST00000398988.3 ENST00000268763.6 |
KSR1
|
kinase suppressor of ras 1 |
| chr19_-_41222775 | 0.12 |
ENST00000324464.3
ENST00000450541.1 ENST00000594720.1 |
ADCK4
|
aarF domain containing kinase 4 |
| chr12_-_7281469 | 0.12 |
ENST00000542370.1
ENST00000266560.3 |
RBP5
|
retinol binding protein 5, cellular |
| chr5_+_118604385 | 0.12 |
ENST00000274456.6
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr11_+_119039069 | 0.12 |
ENST00000422249.1
|
NLRX1
|
NLR family member X1 |
| chr17_+_2264983 | 0.12 |
ENST00000574650.1
|
SGSM2
|
small G protein signaling modulator 2 |
| chr1_-_44818599 | 0.11 |
ENST00000537474.1
|
ERI3
|
ERI1 exoribonuclease family member 3 |
| chr5_+_10564432 | 0.11 |
ENST00000296657.5
|
ANKRD33B
|
ankyrin repeat domain 33B |
| chr1_-_21978312 | 0.11 |
ENST00000359708.4
ENST00000290101.4 |
RAP1GAP
|
RAP1 GTPase activating protein |
| chr17_-_56492989 | 0.11 |
ENST00000583753.1
|
RNF43
|
ring finger protein 43 |
| chr16_-_88717423 | 0.11 |
ENST00000568278.1
ENST00000569359.1 ENST00000567174.1 |
CYBA
|
cytochrome b-245, alpha polypeptide |
| chr2_-_220094031 | 0.11 |
ENST00000443140.1
ENST00000432520.1 ENST00000409618.1 |
ATG9A
|
autophagy related 9A |
| chr19_+_39390587 | 0.11 |
ENST00000572515.1
ENST00000392079.3 ENST00000575359.1 ENST00000313582.5 |
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr1_-_32229523 | 0.11 |
ENST00000398547.1
ENST00000373655.2 ENST00000373658.3 ENST00000257070.4 |
BAI2
|
brain-specific angiogenesis inhibitor 2 |
| chr11_+_65190245 | 0.11 |
ENST00000499732.1
ENST00000501122.2 ENST00000601801.1 |
NEAT1
|
nuclear paraspeckle assembly transcript 1 (non-protein coding) |
| chr19_+_41725140 | 0.11 |
ENST00000359092.3
|
AXL
|
AXL receptor tyrosine kinase |
| chr12_-_49504655 | 0.10 |
ENST00000551782.1
ENST00000267102.8 |
LMBR1L
|
limb development membrane protein 1-like |
| chr6_+_30850697 | 0.10 |
ENST00000509639.1
ENST00000412274.2 ENST00000507901.1 ENST00000507046.1 ENST00000437124.2 ENST00000454612.2 ENST00000396342.2 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr17_-_61920280 | 0.10 |
ENST00000448276.2
ENST00000577990.1 |
SMARCD2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 2 |
| chr1_-_8000872 | 0.10 |
ENST00000377507.3
|
TNFRSF9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr3_-_178789220 | 0.10 |
ENST00000414084.1
|
ZMAT3
|
zinc finger, matrin-type 3 |
| chr6_+_292253 | 0.10 |
ENST00000603453.1
ENST00000605315.1 ENST00000603881.1 |
DUSP22
|
dual specificity phosphatase 22 |
| chr17_-_73511504 | 0.10 |
ENST00000581870.1
|
CASKIN2
|
CASK interacting protein 2 |
| chr2_-_220094294 | 0.10 |
ENST00000436856.1
ENST00000428226.1 ENST00000409422.1 ENST00000431715.1 ENST00000457841.1 ENST00000439812.1 ENST00000361242.4 ENST00000396761.2 |
ATG9A
|
autophagy related 9A |
| chr17_-_76356148 | 0.09 |
ENST00000587578.1
ENST00000330871.2 |
SOCS3
|
suppressor of cytokine signaling 3 |
| chr9_+_132934835 | 0.09 |
ENST00000372398.3
|
NCS1
|
neuronal calcium sensor 1 |
| chr2_+_201987200 | 0.09 |
ENST00000425030.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr2_+_220094479 | 0.09 |
ENST00000323348.5
ENST00000453432.1 ENST00000409849.1 ENST00000416565.1 ENST00000410034.3 ENST00000447157.1 |
ANKZF1
|
ankyrin repeat and zinc finger domain containing 1 |
| chr3_-_183967296 | 0.09 |
ENST00000455059.1
ENST00000445626.2 |
ALG3
|
ALG3, alpha-1,3- mannosyltransferase |
| chr19_+_41222998 | 0.09 |
ENST00000263370.2
|
ITPKC
|
inositol-trisphosphate 3-kinase C |
| chr6_+_32121908 | 0.09 |
ENST00000375143.2
ENST00000424499.1 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr16_+_84209738 | 0.09 |
ENST00000564928.1
|
DNAAF1
|
dynein, axonemal, assembly factor 1 |
| chr2_-_191885686 | 0.08 |
ENST00000432058.1
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr19_-_39390440 | 0.08 |
ENST00000249396.7
ENST00000414941.1 ENST00000392081.2 |
SIRT2
|
sirtuin 2 |
| chr19_-_4831701 | 0.08 |
ENST00000248244.5
|
TICAM1
|
toll-like receptor adaptor molecule 1 |
| chrX_-_153599578 | 0.08 |
ENST00000360319.4
ENST00000344736.4 |
FLNA
|
filamin A, alpha |
| chr22_-_24181174 | 0.08 |
ENST00000318109.7
ENST00000406855.3 ENST00000404056.1 ENST00000476077.1 |
DERL3
|
derlin 3 |
| chr16_-_2059748 | 0.08 |
ENST00000562103.1
ENST00000431526.1 |
ZNF598
|
zinc finger protein 598 |
| chr11_-_910796 | 0.08 |
ENST00000336845.5
ENST00000528581.1 ENST00000525225.1 ENST00000530939.1 ENST00000436108.2 ENST00000429789.2 |
CHID1
|
chitinase domain containing 1 |
| chr12_+_53443680 | 0.08 |
ENST00000314250.6
ENST00000451358.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr3_+_9932238 | 0.08 |
ENST00000307768.4
|
JAGN1
|
jagunal homolog 1 (Drosophila) |
| chr1_-_209824643 | 0.08 |
ENST00000391911.1
ENST00000415782.1 |
LAMB3
|
laminin, beta 3 |
| chr10_+_102756800 | 0.08 |
ENST00000370223.3
|
LZTS2
|
leucine zipper, putative tumor suppressor 2 |
| chr5_-_141703713 | 0.08 |
ENST00000511815.1
|
SPRY4
|
sprouty homolog 4 (Drosophila) |
| chr6_+_30850862 | 0.08 |
ENST00000504651.1
ENST00000512694.1 ENST00000515233.1 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr19_-_39390350 | 0.08 |
ENST00000447739.1
ENST00000358931.5 ENST00000407552.1 |
SIRT2
|
sirtuin 2 |
| chr12_+_53491220 | 0.08 |
ENST00000548547.1
ENST00000301464.3 |
IGFBP6
|
insulin-like growth factor binding protein 6 |
| chr3_+_89156799 | 0.07 |
ENST00000452448.2
ENST00000494014.1 |
EPHA3
|
EPH receptor A3 |
| chr17_-_56591321 | 0.07 |
ENST00000583243.1
|
MTMR4
|
myotubularin related protein 4 |
| chr11_-_75062730 | 0.07 |
ENST00000420843.2
ENST00000360025.3 |
ARRB1
|
arrestin, beta 1 |
| chr4_-_176733897 | 0.07 |
ENST00000393658.2
|
GPM6A
|
glycoprotein M6A |
| chr1_-_231560790 | 0.07 |
ENST00000366641.3
|
EGLN1
|
egl-9 family hypoxia-inducible factor 1 |
| chr6_+_32821924 | 0.07 |
ENST00000374859.2
ENST00000453265.2 |
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr16_-_122619 | 0.07 |
ENST00000262316.6
|
RHBDF1
|
rhomboid 5 homolog 1 (Drosophila) |
| chr22_-_39151434 | 0.07 |
ENST00000439339.1
|
SUN2
|
Sad1 and UNC84 domain containing 2 |
| chr12_-_57504069 | 0.07 |
ENST00000543873.2
ENST00000554663.1 ENST00000557635.1 |
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr17_+_38474489 | 0.07 |
ENST00000394089.2
ENST00000425707.3 |
RARA
|
retinoic acid receptor, alpha |
| chr3_-_53878644 | 0.07 |
ENST00000481668.1
ENST00000467802.1 |
CHDH
|
choline dehydrogenase |
| chr5_+_150591678 | 0.07 |
ENST00000523466.1
|
GM2A
|
GM2 ganglioside activator |
| chr16_+_67571351 | 0.07 |
ENST00000428437.2
ENST00000569253.1 |
FAM65A
|
family with sequence similarity 65, member A |
| chr19_-_41256207 | 0.07 |
ENST00000598485.2
ENST00000470681.1 ENST00000339153.3 ENST00000598729.1 |
C19orf54
|
chromosome 19 open reading frame 54 |
| chr1_-_32110467 | 0.07 |
ENST00000440872.2
ENST00000373703.4 |
PEF1
|
penta-EF-hand domain containing 1 |
| chr3_+_47844615 | 0.07 |
ENST00000348968.4
|
DHX30
|
DEAH (Asp-Glu-Ala-His) box helicase 30 |
| chr2_-_43453734 | 0.06 |
ENST00000282388.3
|
ZFP36L2
|
ZFP36 ring finger protein-like 2 |
| chr5_+_112312416 | 0.06 |
ENST00000389063.2
|
DCP2
|
decapping mRNA 2 |
| chr19_+_13988061 | 0.06 |
ENST00000339133.5
ENST00000397555.2 |
NANOS3
|
nanos homolog 3 (Drosophila) |
| chr1_+_201979645 | 0.06 |
ENST00000367284.5
ENST00000367283.3 |
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr19_-_50370509 | 0.06 |
ENST00000596014.1
|
PNKP
|
polynucleotide kinase 3'-phosphatase |
| chr10_+_104154229 | 0.06 |
ENST00000428099.1
ENST00000369966.3 |
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr16_-_75498553 | 0.06 |
ENST00000569276.1
ENST00000357613.4 ENST00000561878.1 ENST00000566980.1 ENST00000567194.1 |
TMEM170A
RP11-77K12.1
|
transmembrane protein 170A Uncharacterized protein |
| chr16_+_27324983 | 0.06 |
ENST00000566117.1
|
IL4R
|
interleukin 4 receptor |
| chr2_-_114647327 | 0.06 |
ENST00000602760.1
|
RP11-141B14.1
|
RP11-141B14.1 |
| chr15_+_42131011 | 0.06 |
ENST00000458483.1
|
PLA2G4B
|
phospholipase A2, group IVB (cytosolic) |
| chr4_-_103749179 | 0.06 |
ENST00000502690.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr17_-_4643161 | 0.06 |
ENST00000574412.1
|
CXCL16
|
chemokine (C-X-C motif) ligand 16 |
| chr12_+_53443963 | 0.06 |
ENST00000546602.1
ENST00000552570.1 ENST00000549700.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr6_+_32121789 | 0.06 |
ENST00000437001.2
ENST00000375137.2 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr1_+_179050512 | 0.06 |
ENST00000367627.3
|
TOR3A
|
torsin family 3, member A |
| chr1_-_226595648 | 0.06 |
ENST00000366790.3
|
PARP1
|
poly (ADP-ribose) polymerase 1 |
| chr11_-_75062829 | 0.06 |
ENST00000393505.4
|
ARRB1
|
arrestin, beta 1 |
| chr6_-_44233361 | 0.06 |
ENST00000275015.5
|
NFKBIE
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon |
| chr9_+_130547958 | 0.06 |
ENST00000421939.1
ENST00000373265.2 |
CDK9
|
cyclin-dependent kinase 9 |
| chr18_-_56985776 | 0.06 |
ENST00000587244.1
|
CPLX4
|
complexin 4 |
| chr1_+_16767195 | 0.06 |
ENST00000504551.2
ENST00000457722.2 ENST00000406746.1 ENST00000443980.2 |
NECAP2
|
NECAP endocytosis associated 2 |
| chr22_-_24951054 | 0.05 |
ENST00000447813.2
ENST00000402766.1 ENST00000407471.3 ENST00000435822.1 |
GUCD1
|
guanylyl cyclase domain containing 1 |
| chr1_-_32229934 | 0.05 |
ENST00000398542.1
|
BAI2
|
brain-specific angiogenesis inhibitor 2 |
| chr4_-_54930790 | 0.05 |
ENST00000263921.3
|
CHIC2
|
cysteine-rich hydrophobic domain 2 |
| chr20_-_33872518 | 0.05 |
ENST00000374436.3
|
EIF6
|
eukaryotic translation initiation factor 6 |
| chr2_-_74669009 | 0.05 |
ENST00000272430.5
|
RTKN
|
rhotekin |
| chr11_+_65292538 | 0.05 |
ENST00000270176.5
ENST00000525364.1 ENST00000420247.2 ENST00000533862.1 ENST00000279270.6 ENST00000524944.1 |
SCYL1
|
SCY1-like 1 (S. cerevisiae) |
| chr17_+_7590734 | 0.05 |
ENST00000457584.2
|
WRAP53
|
WD repeat containing, antisense to TP53 |
| chr17_-_1420006 | 0.05 |
ENST00000320345.6
ENST00000406424.4 |
INPP5K
|
inositol polyphosphate-5-phosphatase K |
| chr1_-_113249948 | 0.05 |
ENST00000339083.7
ENST00000369642.3 |
RHOC
|
ras homolog family member C |
| chr11_-_64570706 | 0.05 |
ENST00000294066.2
ENST00000377350.3 |
MAP4K2
|
mitogen-activated protein kinase kinase kinase kinase 2 |
| chr20_-_43977055 | 0.05 |
ENST00000372733.3
ENST00000537976.1 |
SDC4
|
syndecan 4 |
| chr19_+_3721719 | 0.05 |
ENST00000589378.1
ENST00000382008.3 |
TJP3
|
tight junction protein 3 |
| chr9_+_130911723 | 0.05 |
ENST00000277480.2
ENST00000373013.2 ENST00000540948.1 |
LCN2
|
lipocalin 2 |
| chr14_-_77227695 | 0.05 |
ENST00000556271.1
|
RP11-99E15.2
|
RP11-99E15.2 |
| chr3_+_9691117 | 0.05 |
ENST00000353332.5
ENST00000420925.1 ENST00000296003.4 ENST00000351233.5 |
MTMR14
|
myotubularin related protein 14 |
| chr20_-_43753104 | 0.05 |
ENST00000372785.3
|
WFDC12
|
WAP four-disulfide core domain 12 |
| chr1_+_56880606 | 0.05 |
ENST00000451914.1
|
RP4-710M16.2
|
RP4-710M16.2 |
| chr6_-_32821599 | 0.05 |
ENST00000354258.4
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr11_-_72492903 | 0.04 |
ENST00000537947.1
|
STARD10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr15_+_44719996 | 0.04 |
ENST00000559793.1
ENST00000558968.1 |
CTDSPL2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr1_-_27816641 | 0.04 |
ENST00000430629.2
|
WASF2
|
WAS protein family, member 2 |
| chr15_-_74753443 | 0.04 |
ENST00000567435.1
ENST00000564488.1 ENST00000565130.1 ENST00000563081.1 ENST00000565335.1 ENST00000395081.2 ENST00000361351.4 |
UBL7
|
ubiquitin-like 7 (bone marrow stromal cell-derived) |
| chr4_+_169418255 | 0.04 |
ENST00000505667.1
ENST00000511948.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr11_+_65292884 | 0.04 |
ENST00000527009.1
|
SCYL1
|
SCY1-like 1 (S. cerevisiae) |
| chr15_+_44719790 | 0.04 |
ENST00000558791.1
|
CTDSPL2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr17_-_1420182 | 0.04 |
ENST00000421807.2
|
INPP5K
|
inositol polyphosphate-5-phosphatase K |
| chr8_+_42128861 | 0.04 |
ENST00000518983.1
|
IKBKB
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta |
| chr19_+_35940486 | 0.04 |
ENST00000246549.2
|
FFAR2
|
free fatty acid receptor 2 |
| chr8_+_123793633 | 0.04 |
ENST00000314393.4
|
ZHX2
|
zinc fingers and homeoboxes 2 |
| chr3_+_57261859 | 0.04 |
ENST00000495803.1
ENST00000444459.1 |
APPL1
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 1 |
| chr9_-_136004782 | 0.04 |
ENST00000393157.3
|
RALGDS
|
ral guanine nucleotide dissociation stimulator |
| chrX_-_134186144 | 0.04 |
ENST00000370775.2
|
FAM127B
|
family with sequence similarity 127, member B |
| chr5_-_127418755 | 0.04 |
ENST00000501702.2
ENST00000501173.2 ENST00000514573.1 ENST00000499346.2 ENST00000606251.1 |
CTC-228N24.3
|
CTC-228N24.3 |
| chr20_-_33872548 | 0.04 |
ENST00000374443.3
|
EIF6
|
eukaryotic translation initiation factor 6 |
| chr2_-_27357479 | 0.04 |
ENST00000406567.3
ENST00000260643.2 |
PREB
|
prolactin regulatory element binding |
| chr5_+_118604439 | 0.04 |
ENST00000388882.5
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr3_-_49142178 | 0.04 |
ENST00000452739.1
ENST00000414533.1 ENST00000417025.1 |
QARS
|
glutaminyl-tRNA synthetase |
| chr16_+_67282853 | 0.04 |
ENST00000299798.11
|
SLC9A5
|
solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 |
| chr8_-_101321584 | 0.04 |
ENST00000523167.1
|
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr1_+_207262170 | 0.04 |
ENST00000367078.3
|
C4BPB
|
complement component 4 binding protein, beta |
| chr4_-_103749205 | 0.04 |
ENST00000508249.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr1_+_165796753 | 0.03 |
ENST00000367879.4
|
UCK2
|
uridine-cytidine kinase 2 |
| chr1_-_204380919 | 0.03 |
ENST00000367188.4
|
PPP1R15B
|
protein phosphatase 1, regulatory subunit 15B |
| chr19_+_14142535 | 0.03 |
ENST00000263379.2
|
IL27RA
|
interleukin 27 receptor, alpha |
| chr5_-_127418573 | 0.03 |
ENST00000508353.1
ENST00000508878.1 ENST00000501652.1 ENST00000514409.1 |
CTC-228N24.3
|
CTC-228N24.3 |
| chr15_+_67430339 | 0.03 |
ENST00000439724.3
|
SMAD3
|
SMAD family member 3 |
| chr19_+_4229495 | 0.03 |
ENST00000221847.5
|
EBI3
|
Epstein-Barr virus induced 3 |
| chr12_-_118490403 | 0.03 |
ENST00000535496.1
|
WSB2
|
WD repeat and SOCS box containing 2 |
| chr19_+_39390320 | 0.03 |
ENST00000576510.1
|
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr7_-_47579188 | 0.03 |
ENST00000398879.1
ENST00000355730.3 ENST00000442536.2 ENST00000458317.2 |
TNS3
|
tensin 3 |
| chr3_-_178789993 | 0.03 |
ENST00000432729.1
|
ZMAT3
|
zinc finger, matrin-type 3 |
| chr12_-_57023995 | 0.03 |
ENST00000549884.1
ENST00000546695.1 |
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A |
| chr11_+_7559485 | 0.03 |
ENST00000527790.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr2_-_61108449 | 0.03 |
ENST00000439412.1
ENST00000452343.1 |
AC010733.4
|
AC010733.4 |
| chr16_+_3096638 | 0.03 |
ENST00000336577.4
|
MMP25
|
matrix metallopeptidase 25 |
| chr8_+_72755367 | 0.03 |
ENST00000537896.1
|
RP11-383H13.1
|
Protein LOC100132891; cDNA FLJ53548 |
| chr17_-_7590745 | 0.03 |
ENST00000514944.1
ENST00000503591.1 ENST00000455263.2 ENST00000420246.2 ENST00000445888.2 ENST00000509690.1 ENST00000604348.1 ENST00000269305.4 |
TP53
|
tumor protein p53 |
| chr19_-_17958832 | 0.03 |
ENST00000458235.1
|
JAK3
|
Janus kinase 3 |
| chr15_+_85923856 | 0.03 |
ENST00000560302.1
ENST00000394518.2 ENST00000361243.2 ENST00000560256.1 |
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr19_+_41725088 | 0.03 |
ENST00000301178.4
|
AXL
|
AXL receptor tyrosine kinase |
| chr19_-_17958771 | 0.03 |
ENST00000534444.1
|
JAK3
|
Janus kinase 3 |
| chr14_-_24616426 | 0.03 |
ENST00000216802.5
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr3_-_112360116 | 0.03 |
ENST00000206423.3
ENST00000439685.2 |
CCDC80
|
coiled-coil domain containing 80 |
| chr7_+_128784712 | 0.03 |
ENST00000289407.4
|
TSPAN33
|
tetraspanin 33 |
| chr5_+_82767284 | 0.03 |
ENST00000265077.3
|
VCAN
|
versican |
| chr1_-_9189144 | 0.03 |
ENST00000414642.2
|
GPR157
|
G protein-coupled receptor 157 |
| chr6_+_12008986 | 0.03 |
ENST00000491710.1
|
HIVEP1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr11_+_73000449 | 0.03 |
ENST00000535931.1
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr12_+_56211703 | 0.03 |
ENST00000243045.5
ENST00000552672.1 ENST00000550836.1 |
ORMDL2
|
ORM1-like 2 (S. cerevisiae) |
| chr14_-_24658053 | 0.03 |
ENST00000354464.6
|
IPO4
|
importin 4 |
| chr7_-_16844611 | 0.03 |
ENST00000401412.1
ENST00000419304.2 |
AGR2
|
anterior gradient 2 |
| chr6_+_15401075 | 0.03 |
ENST00000541660.1
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr7_+_69064300 | 0.03 |
ENST00000342771.4
|
AUTS2
|
autism susceptibility candidate 2 |
| chr15_-_51058005 | 0.03 |
ENST00000261854.5
|
SPPL2A
|
signal peptide peptidase like 2A |
Network of associatons between targets according to the STRING database.
First level regulatory network of REL
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:1902723 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.1 | 0.4 | GO:1901545 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.1 | 0.4 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.1 | GO:0070446 | cellular response to caloric restriction(GO:0061433) negative regulation of oligodendrocyte progenitor proliferation(GO:0070446) |
| 0.0 | 0.1 | GO:2000669 | negative regulation of dendritic cell apoptotic process(GO:2000669) |
| 0.0 | 0.1 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.0 | 0.2 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.4 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.1 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.2 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 0.1 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.0 | 0.1 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.1 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.1 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.1 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.1 | GO:0072308 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.0 | 0.1 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.0 | 0.1 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.0 | 0.2 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.3 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.1 | GO:0033122 | negative regulation of cyclic nucleotide catabolic process(GO:0030806) negative regulation of cAMP catabolic process(GO:0030821) negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 0.0 | 0.1 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.1 | GO:0031456 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.1 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.0 | 0.1 | GO:2001153 | regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 0.0 | 0.1 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.0 | 0.0 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.0 | 0.1 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.2 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.1 | GO:1903971 | mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) positive regulation of microglial cell migration(GO:1904141) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0098502 | DNA dephosphorylation(GO:0098502) |
| 0.0 | 0.0 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.0 | 0.2 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.1 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.0 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.3 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.3 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 0.1 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 0.1 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.1 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0046970 | NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.0 | 0.1 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.3 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0004913 | interleukin-4 receptor activity(GO:0004913) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.1 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.0 | 0.4 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.5 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.1 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID IL27 PATHWAY | IL27-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |