|
chr10_-_38146965
Show fit
|
0.48 |
ENST00000395873.3
ENST00000357328.4
ENST00000395874.2
|
ZNF248
|
zinc finger protein 248
|
|
chr8_-_80680078
Show fit
|
0.42 |
ENST00000337919.5
ENST00000354724.3
|
HEY1
|
hes-related family bHLH transcription factor with YRPW motif 1
|
|
chr1_-_197744763
Show fit
|
0.36 |
ENST00000422998.1
|
DENND1B
|
DENN/MADD domain containing 1B
|
|
chr20_+_32250079
Show fit
|
0.34 |
ENST00000375222.3
|
C20orf144
|
chromosome 20 open reading frame 144
|
|
chr3_-_45957534
Show fit
|
0.34 |
ENST00000536047.1
|
LZTFL1
|
leucine zipper transcription factor-like 1
|
|
chr15_+_59730348
Show fit
|
0.34 |
ENST00000288228.5
ENST00000559628.1
ENST00000557914.1
ENST00000560474.1
|
FAM81A
|
family with sequence similarity 81, member A
|
|
chrX_-_46187069
Show fit
|
0.31 |
ENST00000446884.1
|
RP1-30G7.2
|
RP1-30G7.2
|
|
chr3_-_45957088
Show fit
|
0.30 |
ENST00000539217.1
|
LZTFL1
|
leucine zipper transcription factor-like 1
|
|
chr3_+_32726774
Show fit
|
0.28 |
ENST00000538368.1
|
CNOT10
|
CCR4-NOT transcription complex, subunit 10
|
|
chr6_+_144665237
Show fit
|
0.27 |
ENST00000421035.2
|
UTRN
|
utrophin
|
|
chr2_+_187350973
Show fit
|
0.27 |
ENST00000544130.1
|
ZC3H15
|
zinc finger CCCH-type containing 15
|
|
chr6_-_131299929
Show fit
|
0.24 |
ENST00000531356.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2
|
|
chr3_+_193853927
Show fit
|
0.22 |
ENST00000232424.3
|
HES1
|
hes family bHLH transcription factor 1
|
|
chr16_-_53737722
Show fit
|
0.22 |
ENST00000569716.1
ENST00000562588.1
ENST00000562230.1
ENST00000379925.3
ENST00000563746.1
ENST00000568653.3
|
RPGRIP1L
|
RPGRIP1-like
|
|
chr13_-_74708372
Show fit
|
0.21 |
ENST00000377666.4
|
KLF12
|
Kruppel-like factor 12
|
|
chr2_+_109271481
Show fit
|
0.21 |
ENST00000542845.1
ENST00000393314.2
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chrX_+_106045891
Show fit
|
0.21 |
ENST00000357242.5
ENST00000310452.2
ENST00000481617.2
ENST00000276175.3
|
TBC1D8B
|
TBC1 domain family, member 8B (with GRAM domain)
|
|
chrX_+_16141667
Show fit
|
0.20 |
ENST00000380289.2
|
GRPR
|
gastrin-releasing peptide receptor
|
|
chr10_-_38146510
Show fit
|
0.20 |
ENST00000395867.3
|
ZNF248
|
zinc finger protein 248
|
|
chr11_-_32452357
Show fit
|
0.19 |
ENST00000379079.2
ENST00000530998.1
|
WT1
|
Wilms tumor 1
|
|
chr19_+_19516561
Show fit
|
0.19 |
ENST00000457895.2
|
GATAD2A
|
GATA zinc finger domain containing 2A
|
|
chr6_+_43968306
Show fit
|
0.19 |
ENST00000442114.2
ENST00000336600.5
ENST00000439969.2
|
C6orf223
|
chromosome 6 open reading frame 223
|
|
chr6_+_126070726
Show fit
|
0.18 |
ENST00000368364.3
|
HEY2
|
hes-related family bHLH transcription factor with YRPW motif 2
|
|
chr2_+_187350883
Show fit
|
0.18 |
ENST00000337859.6
|
ZC3H15
|
zinc finger CCCH-type containing 15
|
|
chr19_+_37178482
Show fit
|
0.18 |
ENST00000536254.2
|
ZNF567
|
zinc finger protein 567
|
|
chr3_+_142442841
Show fit
|
0.17 |
ENST00000476941.1
ENST00000273482.6
|
TRPC1
|
transient receptor potential cation channel, subfamily C, member 1
|
|
chr9_+_72658490
Show fit
|
0.17 |
ENST00000377182.4
|
MAMDC2
|
MAM domain containing 2
|
|
chr17_-_16118835
Show fit
|
0.17 |
ENST00000582357.1
ENST00000436828.1
ENST00000411510.1
ENST00000268712.3
|
NCOR1
|
nuclear receptor corepressor 1
|
|
chrX_+_123094369
Show fit
|
0.17 |
ENST00000455404.1
ENST00000218089.9
|
STAG2
|
stromal antigen 2
|
|
chr13_-_25496926
Show fit
|
0.16 |
ENST00000545981.1
ENST00000381884.4
|
CENPJ
|
centromere protein J
|
|
chr19_-_49118067
Show fit
|
0.16 |
ENST00000593772.1
|
FAM83E
|
family with sequence similarity 83, member E
|
|
chr10_-_38146482
Show fit
|
0.16 |
ENST00000374648.3
|
ZNF248
|
zinc finger protein 248
|
|
chr3_+_171561127
Show fit
|
0.16 |
ENST00000334567.5
ENST00000450693.1
|
TMEM212
|
transmembrane protein 212
|
|
chr2_+_44001172
Show fit
|
0.16 |
ENST00000260605.8
ENST00000406852.3
ENST00000443170.3
ENST00000398823.2
ENST00000605786.1
|
DYNC2LI1
|
dynein, cytoplasmic 2, light intermediate chain 1
|
|
chr16_+_4666475
Show fit
|
0.16 |
ENST00000591895.1
|
MGRN1
|
mahogunin ring finger 1, E3 ubiquitin protein ligase
|
|
chr14_+_71788096
Show fit
|
0.16 |
ENST00000557151.1
|
SIPA1L1
|
signal-induced proliferation-associated 1 like 1
|
|
chr10_+_31608054
Show fit
|
0.16 |
ENST00000320985.10
ENST00000361642.5
ENST00000560721.2
ENST00000558440.1
ENST00000424869.1
ENST00000542815.3
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr9_-_98269481
Show fit
|
0.16 |
ENST00000418258.1
ENST00000553011.1
ENST00000551845.1
|
PTCH1
|
patched 1
|
|
chr12_+_6309963
Show fit
|
0.15 |
ENST00000382515.2
|
CD9
|
CD9 molecule
|
|
chr22_+_20862321
Show fit
|
0.15 |
ENST00000541476.1
ENST00000438962.1
|
MED15
|
mediator complex subunit 15
|
|
chr11_-_64512273
Show fit
|
0.15 |
ENST00000377497.3
ENST00000377487.1
ENST00000430645.1
|
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated)
|
|
chr7_-_120498357
Show fit
|
0.15 |
ENST00000415871.1
ENST00000222747.3
ENST00000430985.1
|
TSPAN12
|
tetraspanin 12
|
|
chr3_+_32726620
Show fit
|
0.15 |
ENST00000331889.6
ENST00000328834.5
|
CNOT10
|
CCR4-NOT transcription complex, subunit 10
|
|
chr11_-_64512469
Show fit
|
0.15 |
ENST00000377485.1
|
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated)
|
|
chr12_+_54426637
Show fit
|
0.14 |
ENST00000312492.2
|
HOXC5
|
homeobox C5
|
|
chr1_+_36690011
Show fit
|
0.14 |
ENST00000354618.5
ENST00000469141.2
ENST00000478853.1
|
THRAP3
|
thyroid hormone receptor associated protein 3
|
|
chrX_-_49121165
Show fit
|
0.14 |
ENST00000376207.4
ENST00000376199.2
|
FOXP3
|
forkhead box P3
|
|
chr6_-_76203345
Show fit
|
0.14 |
ENST00000393004.2
|
FILIP1
|
filamin A interacting protein 1
|
|
chr2_-_111230393
Show fit
|
0.14 |
ENST00000447537.2
ENST00000413601.2
|
LIMS3L
LIMS3L
|
LIM and senescent cell antigen-like-containing domain protein 3; Uncharacterized protein; cDNA FLJ59124, highly similar to Particularly interesting newCys-His protein; cDNA, FLJ79109, highly similar to Particularly interesting newCys-His protein
LIM and senescent cell antigen-like domains 3-like
|
|
chr17_-_62499334
Show fit
|
0.14 |
ENST00000579996.1
|
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5
|
|
chr2_+_208104497
Show fit
|
0.14 |
ENST00000430494.1
|
AC007879.7
|
AC007879.7
|
|
chr1_+_239882842
Show fit
|
0.14 |
ENST00000448020.1
|
CHRM3
|
cholinergic receptor, muscarinic 3
|
|
chr17_+_29421987
Show fit
|
0.13 |
ENST00000431387.4
|
NF1
|
neurofibromin 1
|
|
chr11_+_64053005
Show fit
|
0.13 |
ENST00000538032.1
|
GPR137
|
G protein-coupled receptor 137
|
|
chr14_-_53258180
Show fit
|
0.13 |
ENST00000554230.1
|
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1
|
|
chr16_-_30102547
Show fit
|
0.13 |
ENST00000279386.2
|
TBX6
|
T-box 6
|
|
chr3_+_159570722
Show fit
|
0.13 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr16_-_53737795
Show fit
|
0.13 |
ENST00000262135.4
ENST00000564374.1
ENST00000566096.1
|
RPGRIP1L
|
RPGRIP1-like
|
|
chr1_-_223537401
Show fit
|
0.13 |
ENST00000343846.3
ENST00000454695.2
ENST00000484758.2
|
SUSD4
|
sushi domain containing 4
|
|
chr10_+_112836779
Show fit
|
0.13 |
ENST00000280155.2
|
ADRA2A
|
adrenoceptor alpha 2A
|
|
chrX_-_153095813
Show fit
|
0.13 |
ENST00000544474.1
|
PDZD4
|
PDZ domain containing 4
|
|
chr22_+_20861858
Show fit
|
0.13 |
ENST00000414658.1
ENST00000432052.1
ENST00000425759.2
ENST00000292733.7
ENST00000542773.1
ENST00000263205.7
ENST00000406969.1
ENST00000382974.2
|
MED15
|
mediator complex subunit 15
|
|
chrX_+_19362011
Show fit
|
0.12 |
ENST00000379806.5
ENST00000545074.1
ENST00000540249.1
ENST00000423505.1
ENST00000417819.1
ENST00000422285.2
ENST00000355808.5
ENST00000379805.3
|
PDHA1
|
pyruvate dehydrogenase (lipoamide) alpha 1
|
|
chr1_-_243349684
Show fit
|
0.12 |
ENST00000522895.1
|
CEP170
|
centrosomal protein 170kDa
|
|
chr17_+_29421900
Show fit
|
0.12 |
ENST00000358273.4
ENST00000356175.3
|
NF1
|
neurofibromin 1
|
|
chr9_+_102584128
Show fit
|
0.12 |
ENST00000338488.4
ENST00000395097.2
|
NR4A3
|
nuclear receptor subfamily 4, group A, member 3
|
|
chr1_+_93297582
Show fit
|
0.12 |
ENST00000370321.3
|
RPL5
|
ribosomal protein L5
|
|
chr14_-_53417732
Show fit
|
0.12 |
ENST00000399304.3
ENST00000395631.2
ENST00000341590.3
ENST00000343279.4
|
FERMT2
|
fermitin family member 2
|
|
chr10_-_74927810
Show fit
|
0.12 |
ENST00000372979.4
ENST00000430082.2
ENST00000454759.2
ENST00000413026.1
ENST00000453402.1
|
ECD
|
ecdysoneless homolog (Drosophila)
|
|
chr3_+_107241882
Show fit
|
0.11 |
ENST00000416476.2
|
BBX
|
bobby sox homolog (Drosophila)
|
|
chr7_-_27169801
Show fit
|
0.11 |
ENST00000511914.1
|
HOXA4
|
homeobox A4
|
|
chr1_-_146644036
Show fit
|
0.11 |
ENST00000425272.2
|
PRKAB2
|
protein kinase, AMP-activated, beta 2 non-catalytic subunit
|
|
chrX_+_123095546
Show fit
|
0.11 |
ENST00000371157.3
ENST00000371145.3
ENST00000371144.3
|
STAG2
|
stromal antigen 2
|
|
chr16_+_75182376
Show fit
|
0.11 |
ENST00000570010.1
ENST00000568079.1
ENST00000464850.1
ENST00000332307.4
ENST00000393430.2
|
ZFP1
|
ZFP1 zinc finger protein
|
|
chr18_+_10526008
Show fit
|
0.11 |
ENST00000542979.1
ENST00000322897.6
|
NAPG
|
N-ethylmaleimide-sensitive factor attachment protein, gamma
|
|
chr1_-_149459549
Show fit
|
0.10 |
ENST00000369175.3
|
FAM72C
|
family with sequence similarity 72, member C
|
|
chr10_+_124320156
Show fit
|
0.10 |
ENST00000338354.3
ENST00000344338.3
ENST00000330163.4
ENST00000368909.3
ENST00000368955.3
ENST00000368956.2
|
DMBT1
|
deleted in malignant brain tumors 1
|
|
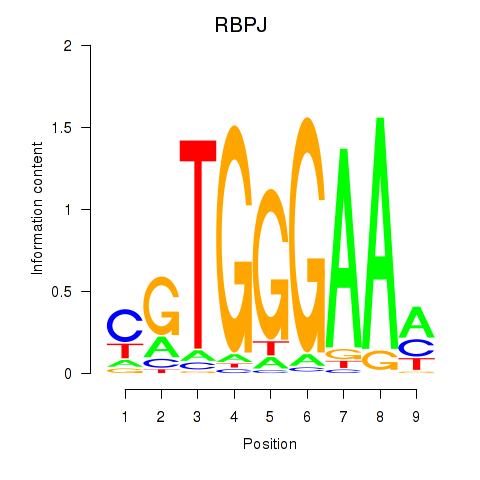
chr4_+_26321284
Show fit
|
0.10 |
ENST00000506956.1
ENST00000512671.1
ENST00000345843.3
ENST00000342295.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region
|
|
chr11_+_45944190
Show fit
|
0.10 |
ENST00000401752.1
ENST00000389968.3
ENST00000325468.5
ENST00000536139.1
|
GYLTL1B
|
glycosyltransferase-like 1B
|
|
chr1_-_197744390
Show fit
|
0.10 |
ENST00000367396.3
|
DENND1B
|
DENN/MADD domain containing 1B
|
|
chrX_-_118739835
Show fit
|
0.10 |
ENST00000542113.1
ENST00000304449.5
|
NKRF
|
NFKB repressing factor
|
|
chr2_+_183580954
Show fit
|
0.10 |
ENST00000264065.7
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10
|
|
chr14_-_24701539
Show fit
|
0.10 |
ENST00000534348.1
ENST00000524927.1
ENST00000250495.5
|
NEDD8-MDP1
NEDD8
|
NEDD8-MDP1 readthrough
neural precursor cell expressed, developmentally down-regulated 8
|
|
chr10_-_27530997
Show fit
|
0.10 |
ENST00000375901.1
ENST00000412279.1
ENST00000375905.4
|
ACBD5
|
acyl-CoA binding domain containing 5
|
|
chr19_+_36346734
Show fit
|
0.09 |
ENST00000586102.3
|
KIRREL2
|
kin of IRRE like 2 (Drosophila)
|
|
chr6_-_72129806
Show fit
|
0.09 |
ENST00000413945.1
ENST00000602878.1
ENST00000436803.1
ENST00000421704.1
ENST00000441570.1
|
LINC00472
|
long intergenic non-protein coding RNA 472
|
|
chr2_+_110656268
Show fit
|
0.09 |
ENST00000553749.1
|
LIMS3
|
LIM and senescent cell antigen-like domains 3
|
|
chr12_+_4918342
Show fit
|
0.09 |
ENST00000280684.3
ENST00000433855.1
|
KCNA6
|
potassium voltage-gated channel, shaker-related subfamily, member 6
|
|
chr6_-_94129244
Show fit
|
0.09 |
ENST00000369303.4
ENST00000369297.1
|
EPHA7
|
EPH receptor A7
|
|
chr6_+_20403997
Show fit
|
0.08 |
ENST00000535432.1
|
E2F3
|
E2F transcription factor 3
|
|
chr1_+_185126291
Show fit
|
0.08 |
ENST00000367500.4
|
SWT1
|
SWT1 RNA endoribonuclease homolog (S. cerevisiae)
|
|
chr17_-_56606664
Show fit
|
0.08 |
ENST00000580844.1
|
SEPT4
|
septin 4
|
|
chr12_+_56623827
Show fit
|
0.08 |
ENST00000424625.1
ENST00000419753.1
ENST00000454355.2
ENST00000417965.1
ENST00000436633.1
|
SLC39A5
|
solute carrier family 39 (zinc transporter), member 5
|
|
chr11_-_107436443
Show fit
|
0.08 |
ENST00000429370.1
ENST00000417449.2
ENST00000428149.2
|
ALKBH8
|
alkB, alkylation repair homolog 8 (E. coli)
|
|
chr16_+_53738053
Show fit
|
0.08 |
ENST00000394647.3
|
FTO
|
fat mass and obesity associated
|
|
chr13_-_38443860
Show fit
|
0.07 |
ENST00000426868.2
ENST00000379681.3
ENST00000338947.5
ENST00000355779.2
ENST00000358477.2
ENST00000379673.2
|
TRPC4
|
transient receptor potential cation channel, subfamily C, member 4
|
|
chr12_-_31945172
Show fit
|
0.07 |
ENST00000340398.3
|
H3F3C
|
H3 histone, family 3C
|
|
chr1_+_82165350
Show fit
|
0.07 |
ENST00000359929.3
|
LPHN2
|
latrophilin 2
|
|
chr5_+_137514687
Show fit
|
0.07 |
ENST00000394894.3
|
KIF20A
|
kinesin family member 20A
|
|
chr17_-_2239729
Show fit
|
0.07 |
ENST00000576112.2
|
TSR1
|
TSR1, 20S rRNA accumulation, homolog (S. cerevisiae)
|
|
chr9_-_98268883
Show fit
|
0.07 |
ENST00000551630.1
ENST00000548420.1
|
PTCH1
|
patched 1
|
|
chr9_+_88556036
Show fit
|
0.07 |
ENST00000361671.5
ENST00000416045.1
|
NAA35
|
N(alpha)-acetyltransferase 35, NatC auxiliary subunit
|
|
chr5_-_39074479
Show fit
|
0.07 |
ENST00000514735.1
ENST00000296782.5
ENST00000357387.3
|
RICTOR
|
RPTOR independent companion of MTOR, complex 2
|
|
chr11_-_64512803
Show fit
|
0.07 |
ENST00000377489.1
ENST00000354024.3
|
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated)
|
|
chr10_+_60144782
Show fit
|
0.07 |
ENST00000487519.1
|
TFAM
|
transcription factor A, mitochondrial
|
|
chr22_-_31885727
Show fit
|
0.07 |
ENST00000330125.5
ENST00000344710.5
ENST00000397518.1
|
EIF4ENIF1
|
eukaryotic translation initiation factor 4E nuclear import factor 1
|
|
chr11_-_101454658
Show fit
|
0.07 |
ENST00000344327.3
|
TRPC6
|
transient receptor potential cation channel, subfamily C, member 6
|
|
chr19_+_50529212
Show fit
|
0.07 |
ENST00000270617.3
ENST00000445728.3
ENST00000601364.1
|
ZNF473
|
zinc finger protein 473
|
|
chr7_-_94953878
Show fit
|
0.07 |
ENST00000222381.3
|
PON1
|
paraoxonase 1
|
|
chr4_-_164534657
Show fit
|
0.06 |
ENST00000339875.5
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase
|
|
chr17_-_56606705
Show fit
|
0.06 |
ENST00000317268.3
|
SEPT4
|
septin 4
|
|
chr1_+_15272271
Show fit
|
0.06 |
ENST00000400797.3
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr16_-_10276251
Show fit
|
0.06 |
ENST00000330684.3
|
GRIN2A
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A
|
|
chr1_-_223536679
Show fit
|
0.06 |
ENST00000608996.1
|
SUSD4
|
sushi domain containing 4
|
|
chrX_-_11369656
Show fit
|
0.06 |
ENST00000413512.3
|
ARHGAP6
|
Rho GTPase activating protein 6
|
|
chr5_+_137514834
Show fit
|
0.06 |
ENST00000508792.1
ENST00000504621.1
|
KIF20A
|
kinesin family member 20A
|
|
chr1_-_935491
Show fit
|
0.06 |
ENST00000304952.6
|
HES4
|
hes family bHLH transcription factor 4
|
|
chr15_-_33486897
Show fit
|
0.06 |
ENST00000320930.7
|
FMN1
|
formin 1
|
|
chr5_+_63461642
Show fit
|
0.06 |
ENST00000296615.6
ENST00000381081.2
ENST00000389100.4
|
RNF180
|
ring finger protein 180
|
|
chr10_+_74927875
Show fit
|
0.06 |
ENST00000242505.6
|
FAM149B1
|
family with sequence similarity 149, member B1
|
|
chr2_+_106361333
Show fit
|
0.06 |
ENST00000233154.4
ENST00000451463.2
|
NCK2
|
NCK adaptor protein 2
|
|
chr14_+_51955831
Show fit
|
0.06 |
ENST00000356218.4
|
FRMD6
|
FERM domain containing 6
|
|
chr11_+_66512089
Show fit
|
0.06 |
ENST00000524551.1
ENST00000525908.1
ENST00000360962.4
ENST00000346672.4
ENST00000527634.1
ENST00000540737.1
|
C11orf80
|
chromosome 11 open reading frame 80
|
|
chr10_+_124320195
Show fit
|
0.06 |
ENST00000359586.6
|
DMBT1
|
deleted in malignant brain tumors 1
|
|
chr1_-_211752073
Show fit
|
0.06 |
ENST00000367001.4
|
SLC30A1
|
solute carrier family 30 (zinc transporter), member 1
|
|
chr11_+_66512303
Show fit
|
0.06 |
ENST00000532565.2
|
C11orf80
|
chromosome 11 open reading frame 80
|
|
chr5_-_137514333
Show fit
|
0.06 |
ENST00000411594.2
ENST00000430331.1
|
BRD8
|
bromodomain containing 8
|
|
chr19_+_5681153
Show fit
|
0.06 |
ENST00000579559.1
ENST00000577222.1
|
HSD11B1L
RPL36
|
hydroxysteroid (11-beta) dehydrogenase 1-like
ribosomal protein L36
|
|
chr1_-_180471947
Show fit
|
0.05 |
ENST00000367595.3
|
ACBD6
|
acyl-CoA binding domain containing 6
|
|
chr19_+_50528971
Show fit
|
0.05 |
ENST00000598809.1
ENST00000595661.1
ENST00000391821.2
|
ZNF473
|
zinc finger protein 473
|
|
chr5_-_137514288
Show fit
|
0.05 |
ENST00000454473.1
ENST00000418329.1
ENST00000455658.2
ENST00000230901.5
ENST00000402931.1
|
BRD8
|
bromodomain containing 8
|
|
chr13_-_39612176
Show fit
|
0.05 |
ENST00000352251.3
ENST00000350125.3
|
PROSER1
|
proline and serine rich 1
|
|
chr11_+_7598239
Show fit
|
0.05 |
ENST00000525597.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2)
|
|
chr6_+_42531798
Show fit
|
0.05 |
ENST00000372903.2
ENST00000372899.1
ENST00000372901.1
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2
|
|
chr11_+_112832133
Show fit
|
0.05 |
ENST00000524665.1
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr22_-_38245304
Show fit
|
0.05 |
ENST00000609454.1
|
ANKRD54
|
ankyrin repeat domain 54
|
|
chr10_-_97200772
Show fit
|
0.05 |
ENST00000371241.1
ENST00000354106.3
ENST00000371239.1
ENST00000361941.3
ENST00000277982.5
ENST00000371245.3
|
SORBS1
|
sorbin and SH3 domain containing 1
|
|
chr11_+_112832202
Show fit
|
0.05 |
ENST00000534015.1
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr17_-_56606639
Show fit
|
0.05 |
ENST00000579371.1
|
SEPT4
|
septin 4
|
|
chr2_-_170219037
Show fit
|
0.05 |
ENST00000443831.1
|
LRP2
|
low density lipoprotein receptor-related protein 2
|
|
chr13_-_39611956
Show fit
|
0.05 |
ENST00000418503.1
|
PROSER1
|
proline and serine rich 1
|
|
chr2_+_208104351
Show fit
|
0.04 |
ENST00000440326.1
|
AC007879.7
|
AC007879.7
|
|
chr20_+_2633269
Show fit
|
0.04 |
ENST00000445139.1
|
NOP56
|
NOP56 ribonucleoprotein
|
|
chr2_+_204193101
Show fit
|
0.04 |
ENST00000430418.1
ENST00000424558.1
ENST00000261016.6
|
ABI2
|
abl-interactor 2
|
|
chr1_+_113933371
Show fit
|
0.04 |
ENST00000369617.4
|
MAGI3
|
membrane associated guanylate kinase, WW and PDZ domain containing 3
|
|
chrX_+_123095890
Show fit
|
0.04 |
ENST00000435215.1
|
STAG2
|
stromal antigen 2
|
|
chrX_-_153095945
Show fit
|
0.04 |
ENST00000164640.4
|
PDZD4
|
PDZ domain containing 4
|
|
chr3_-_171489085
Show fit
|
0.04 |
ENST00000418087.1
|
PLD1
|
phospholipase D1, phosphatidylcholine-specific
|
|
chr1_-_235667716
Show fit
|
0.04 |
ENST00000313984.3
ENST00000366600.3
|
B3GALNT2
|
beta-1,3-N-acetylgalactosaminyltransferase 2
|
|
chr1_+_210406121
Show fit
|
0.04 |
ENST00000367012.3
|
SERTAD4
|
SERTA domain containing 4
|
|
chr14_+_73603126
Show fit
|
0.04 |
ENST00000557356.1
ENST00000556864.1
ENST00000556533.1
ENST00000556951.1
ENST00000557293.1
ENST00000553719.1
ENST00000553599.1
ENST00000556011.1
ENST00000394157.3
ENST00000357710.4
ENST00000324501.5
ENST00000560005.2
ENST00000555254.1
ENST00000261970.3
ENST00000344094.3
ENST00000554131.1
ENST00000557037.1
|
PSEN1
|
presenilin 1
|
|
chr3_-_119813264
Show fit
|
0.04 |
ENST00000264235.8
|
GSK3B
|
glycogen synthase kinase 3 beta
|
|
chr18_+_66382428
Show fit
|
0.04 |
ENST00000578970.1
ENST00000582371.1
ENST00000584775.1
|
CCDC102B
|
coiled-coil domain containing 102B
|
|
chr9_+_706842
Show fit
|
0.04 |
ENST00000382293.3
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr9_-_35691017
Show fit
|
0.04 |
ENST00000378292.3
|
TPM2
|
tropomyosin 2 (beta)
|
|
chr11_-_57102947
Show fit
|
0.04 |
ENST00000526696.1
|
SSRP1
|
structure specific recognition protein 1
|
|
chr5_-_175815565
Show fit
|
0.04 |
ENST00000509257.1
ENST00000507413.1
ENST00000510123.1
|
NOP16
|
NOP16 nucleolar protein
|
|
chr6_-_76203454
Show fit
|
0.04 |
ENST00000237172.7
|
FILIP1
|
filamin A interacting protein 1
|
|
chr14_-_93214988
Show fit
|
0.04 |
ENST00000557434.1
ENST00000393218.2
ENST00000334869.4
|
LGMN
|
legumain
|
|
chr3_+_107241783
Show fit
|
0.04 |
ENST00000415149.2
ENST00000402543.1
ENST00000325805.8
ENST00000427402.1
|
BBX
|
bobby sox homolog (Drosophila)
|
|
chrX_-_118925600
Show fit
|
0.04 |
ENST00000361575.3
|
RPL39
|
ribosomal protein L39
|
|
chr15_+_75182346
Show fit
|
0.04 |
ENST00000569931.1
ENST00000352410.4
ENST00000566377.1
ENST00000569233.1
ENST00000567132.1
ENST00000564633.1
ENST00000568907.1
ENST00000563422.1
ENST00000564003.1
ENST00000562800.1
ENST00000563786.1
ENST00000535694.1
ENST00000323744.6
ENST00000568828.1
ENST00000562606.1
ENST00000565576.1
ENST00000567570.1
|
MPI
|
mannose phosphate isomerase
|
|
chr7_+_30811004
Show fit
|
0.04 |
ENST00000265299.6
|
FAM188B
|
family with sequence similarity 188, member B
|
|
chr1_+_61547405
Show fit
|
0.04 |
ENST00000371189.4
|
NFIA
|
nuclear factor I/A
|
|
chr13_-_20357057
Show fit
|
0.04 |
ENST00000338910.4
|
PSPC1
|
paraspeckle component 1
|
|
chr8_-_67974552
Show fit
|
0.04 |
ENST00000357849.4
|
COPS5
|
COP9 signalosome subunit 5
|
|
chr16_-_70719925
Show fit
|
0.04 |
ENST00000338779.6
|
MTSS1L
|
metastasis suppressor 1-like
|
|
chr19_-_50529193
Show fit
|
0.04 |
ENST00000596445.1
ENST00000599538.1
|
VRK3
|
vaccinia related kinase 3
|
|
chr21_+_38792602
Show fit
|
0.04 |
ENST00000398960.2
ENST00000398956.2
|
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A
|
|
chr17_+_41322483
Show fit
|
0.03 |
ENST00000341165.6
ENST00000586650.1
ENST00000422280.1
|
NBR1
|
neighbor of BRCA1 gene 1
|
|
chr5_-_137514617
Show fit
|
0.03 |
ENST00000254900.5
|
BRD8
|
bromodomain containing 8
|
|
chr5_+_178368186
Show fit
|
0.03 |
ENST00000320129.3
ENST00000519564.1
|
ZNF454
|
zinc finger protein 454
|
|
chr16_+_72459838
Show fit
|
0.03 |
ENST00000564508.1
|
AC004158.3
|
AC004158.3
|
|
chr10_+_118305435
Show fit
|
0.03 |
ENST00000369221.2
|
PNLIP
|
pancreatic lipase
|
|
chr3_-_100712292
Show fit
|
0.03 |
ENST00000495063.1
ENST00000530539.1
|
ABI3BP
|
ABI family, member 3 (NESH) binding protein
|
|
chr1_+_110026544
Show fit
|
0.03 |
ENST00000369870.3
|
ATXN7L2
|
ataxin 7-like 2
|
|
chr19_-_39390212
Show fit
|
0.03 |
ENST00000437828.1
|
SIRT2
|
sirtuin 2
|
|
chr1_-_197036364
Show fit
|
0.03 |
ENST00000367412.1
|
F13B
|
coagulation factor XIII, B polypeptide
|
|
chr16_-_10276611
Show fit
|
0.03 |
ENST00000396573.2
|
GRIN2A
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A
|
|
chr7_+_102553430
Show fit
|
0.03 |
ENST00000339431.4
ENST00000249377.4
|
LRRC17
|
leucine rich repeat containing 17
|
|
chr11_+_112832090
Show fit
|
0.03 |
ENST00000533760.1
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr11_-_8959758
Show fit
|
0.03 |
ENST00000531618.1
|
ASCL3
|
achaete-scute family bHLH transcription factor 3
|
|
chrX_+_123095860
Show fit
|
0.03 |
ENST00000428941.1
|
STAG2
|
stromal antigen 2
|
|
chr5_-_137071689
Show fit
|
0.03 |
ENST00000505853.1
|
KLHL3
|
kelch-like family member 3
|
|
chr10_-_44144152
Show fit
|
0.03 |
ENST00000395797.1
|
ZNF32
|
zinc finger protein 32
|
|
chr10_+_115439699
Show fit
|
0.03 |
ENST00000369315.1
|
CASP7
|
caspase 7, apoptosis-related cysteine peptidase
|
|
chr1_+_8021954
Show fit
|
0.03 |
ENST00000377491.1
ENST00000377488.1
|
PARK7
|
parkinson protein 7
|
|
chr7_+_55177416
Show fit
|
0.03 |
ENST00000450046.1
ENST00000454757.2
|
EGFR
|
epidermal growth factor receptor
|
|
chr19_-_36870087
Show fit
|
0.03 |
ENST00000270001.7
|
ZFP14
|
ZFP14 zinc finger protein
|
|
chr2_-_164592497
Show fit
|
0.03 |
ENST00000333129.3
ENST00000409634.1
|
FIGN
|
fidgetin
|
|
chr3_+_184016986
Show fit
|
0.03 |
ENST00000417952.1
|
PSMD2
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 2
|
|
chr7_-_137531606
Show fit
|
0.03 |
ENST00000288490.5
|
DGKI
|
diacylglycerol kinase, iota
|
|
chr10_-_33623826
Show fit
|
0.02 |
ENST00000374867.2
|
NRP1
|
neuropilin 1
|
|
chrX_-_73834449
Show fit
|
0.02 |
ENST00000332687.6
ENST00000349225.2
|
RLIM
|
ring finger protein, LIM domain interacting
|
|
chr14_+_29234870
Show fit
|
0.02 |
ENST00000382535.3
|
FOXG1
|
forkhead box G1
|
|
chr14_-_48144157
Show fit
|
0.02 |
ENST00000399232.2
|
MDGA2
|
MAM domain containing glycosylphosphatidylinositol anchor 2
|
|
chr12_-_90049878
Show fit
|
0.02 |
ENST00000359142.3
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1
|
|
chr10_-_49459800
Show fit
|
0.02 |
ENST00000305531.3
|
FRMPD2
|
FERM and PDZ domain containing 2
|
|
chr7_-_16505440
Show fit
|
0.02 |
ENST00000307068.4
|
SOSTDC1
|
sclerostin domain containing 1
|
|
chr5_-_124080203
Show fit
|
0.02 |
ENST00000504926.1
|
ZNF608
|
zinc finger protein 608
|