Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
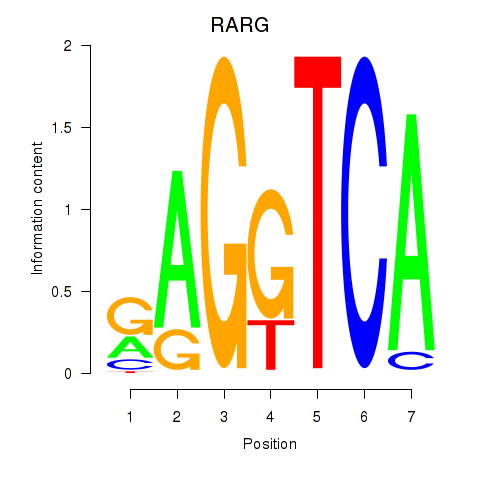
Results for RARG
Z-value: 0.31
Transcription factors associated with RARG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RARG
|
ENSG00000172819.12 | retinoic acid receptor gamma |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RARG | hg19_v2_chr12_-_53625958_53626036 | -0.77 | 2.3e-01 | Click! |
Activity profile of RARG motif
Sorted Z-values of RARG motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_117716418 | 0.19 |
ENST00000484092.1
|
RP11-384F7.2
|
RP11-384F7.2 |
| chr19_+_49467232 | 0.12 |
ENST00000599784.1
ENST00000594305.1 |
CTD-2639E6.9
|
CTD-2639E6.9 |
| chr19_-_51289374 | 0.12 |
ENST00000563228.1
|
CTD-2568A17.1
|
CTD-2568A17.1 |
| chr6_-_30080863 | 0.11 |
ENST00000540829.1
|
TRIM31
|
tripartite motif containing 31 |
| chr8_-_131028660 | 0.10 |
ENST00000401979.2
ENST00000517654.1 ENST00000522361.1 ENST00000518167.1 |
FAM49B
|
family with sequence similarity 49, member B |
| chr1_-_17380630 | 0.10 |
ENST00000375499.3
|
SDHB
|
succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
| chr2_-_219433014 | 0.10 |
ENST00000418019.1
ENST00000454775.1 ENST00000338465.5 ENST00000415516.1 ENST00000258399.3 |
USP37
|
ubiquitin specific peptidase 37 |
| chr9_-_34381536 | 0.10 |
ENST00000379126.3
ENST00000379127.1 ENST00000379133.3 |
C9orf24
|
chromosome 9 open reading frame 24 |
| chr17_-_49021974 | 0.09 |
ENST00000501718.2
|
RP11-700H6.1
|
RP11-700H6.1 |
| chr9_-_88896977 | 0.09 |
ENST00000311534.6
|
ISCA1
|
iron-sulfur cluster assembly 1 |
| chr19_-_51568324 | 0.09 |
ENST00000595547.1
ENST00000335422.3 ENST00000595793.1 ENST00000596955.1 |
KLK13
|
kallikrein-related peptidase 13 |
| chr9_-_16705069 | 0.08 |
ENST00000471301.2
|
BNC2
|
basonuclin 2 |
| chr8_-_131028869 | 0.08 |
ENST00000518283.1
ENST00000519110.1 |
FAM49B
|
family with sequence similarity 49, member B |
| chr10_+_81065975 | 0.08 |
ENST00000446377.2
|
ZMIZ1
|
zinc finger, MIZ-type containing 1 |
| chr5_-_87564620 | 0.08 |
ENST00000506536.1
ENST00000512429.1 ENST00000514135.1 ENST00000296595.6 ENST00000509387.1 |
TMEM161B
|
transmembrane protein 161B |
| chr15_+_65337708 | 0.08 |
ENST00000334287.2
|
SLC51B
|
solute carrier family 51, beta subunit |
| chr8_+_38261880 | 0.08 |
ENST00000527175.1
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr19_-_51289436 | 0.08 |
ENST00000562076.1
|
CTD-2568A17.1
|
CTD-2568A17.1 |
| chr12_-_16760195 | 0.08 |
ENST00000546281.1
ENST00000537757.1 |
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr21_-_27107198 | 0.08 |
ENST00000400094.1
|
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr3_+_37284668 | 0.07 |
ENST00000361924.2
ENST00000444882.1 ENST00000356847.4 ENST00000450863.2 ENST00000429018.1 |
GOLGA4
|
golgin A4 |
| chr1_-_150693318 | 0.07 |
ENST00000442853.1
ENST00000368995.4 ENST00000368993.2 ENST00000361824.2 ENST00000322343.7 |
HORMAD1
|
HORMA domain containing 1 |
| chr6_-_30080876 | 0.07 |
ENST00000376734.3
|
TRIM31
|
tripartite motif containing 31 |
| chrX_-_46187069 | 0.07 |
ENST00000446884.1
|
RP1-30G7.2
|
RP1-30G7.2 |
| chr1_+_29213678 | 0.07 |
ENST00000347529.3
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
| chr4_+_106067943 | 0.07 |
ENST00000380013.4
ENST00000394764.1 ENST00000413648.2 |
TET2
|
tet methylcytosine dioxygenase 2 |
| chr9_+_139873264 | 0.07 |
ENST00000446677.1
|
PTGDS
|
prostaglandin D2 synthase 21kDa (brain) |
| chr19_-_23869999 | 0.07 |
ENST00000601935.1
ENST00000359788.4 ENST00000600313.1 ENST00000596211.1 ENST00000599168.1 |
ZNF675
|
zinc finger protein 675 |
| chr16_+_222846 | 0.07 |
ENST00000251595.6
ENST00000397806.1 |
HBA2
|
hemoglobin, alpha 2 |
| chr15_-_71184724 | 0.07 |
ENST00000560604.1
|
THAP10
|
THAP domain containing 10 |
| chr3_-_45957088 | 0.07 |
ENST00000539217.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr6_-_134861089 | 0.07 |
ENST00000606039.1
|
RP11-557H15.4
|
RP11-557H15.4 |
| chr16_-_11477562 | 0.07 |
ENST00000595168.1
|
CTD-3088G3.8
|
Protein LOC388210 |
| chr14_-_21492251 | 0.07 |
ENST00000554398.1
|
NDRG2
|
NDRG family member 2 |
| chr1_-_245134273 | 0.07 |
ENST00000607453.1
|
RP11-156E8.1
|
Uncharacterized protein |
| chr11_+_63974135 | 0.07 |
ENST00000544997.1
ENST00000345728.5 ENST00000279227.5 |
FERMT3
|
fermitin family member 3 |
| chr17_+_7533439 | 0.07 |
ENST00000441599.2
ENST00000380450.4 ENST00000416273.3 ENST00000575903.1 ENST00000576830.1 ENST00000571153.1 ENST00000575618.1 ENST00000576152.1 |
SHBG
|
sex hormone-binding globulin |
| chr19_-_49371711 | 0.07 |
ENST00000355496.5
ENST00000263265.6 |
PLEKHA4
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 4 |
| chr1_-_85156417 | 0.07 |
ENST00000422026.1
|
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr17_-_79792909 | 0.06 |
ENST00000330261.4
ENST00000570394.1 |
PPP1R27
|
protein phosphatase 1, regulatory subunit 27 |
| chr1_+_178482262 | 0.06 |
ENST00000367641.3
ENST00000367639.1 |
TEX35
|
testis expressed 35 |
| chr2_+_219283815 | 0.06 |
ENST00000248444.5
ENST00000454069.1 ENST00000392114.2 |
VIL1
|
villin 1 |
| chr15_+_63414760 | 0.06 |
ENST00000557972.1
|
LACTB
|
lactamase, beta |
| chr2_-_192016276 | 0.06 |
ENST00000413064.1
|
STAT4
|
signal transducer and activator of transcription 4 |
| chr20_-_57617831 | 0.06 |
ENST00000371033.5
ENST00000355937.4 |
SLMO2
|
slowmo homolog 2 (Drosophila) |
| chr17_+_8316442 | 0.06 |
ENST00000582812.1
|
NDEL1
|
nudE neurodevelopment protein 1-like 1 |
| chr9_-_34381511 | 0.06 |
ENST00000379124.1
|
C9orf24
|
chromosome 9 open reading frame 24 |
| chr12_+_57854274 | 0.06 |
ENST00000528432.1
|
GLI1
|
GLI family zinc finger 1 |
| chr1_+_112016414 | 0.06 |
ENST00000343534.5
ENST00000369718.3 |
C1orf162
|
chromosome 1 open reading frame 162 |
| chr1_+_222988464 | 0.06 |
ENST00000420335.1
|
RP11-452F19.3
|
RP11-452F19.3 |
| chr7_+_79764104 | 0.06 |
ENST00000351004.3
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr3_-_52486841 | 0.06 |
ENST00000496590.1
|
TNNC1
|
troponin C type 1 (slow) |
| chr12_+_123874589 | 0.06 |
ENST00000437502.1
|
SETD8
|
SET domain containing (lysine methyltransferase) 8 |
| chr19_+_46531127 | 0.06 |
ENST00000601033.1
|
CTC-344H19.4
|
CTC-344H19.4 |
| chr2_+_33701684 | 0.06 |
ENST00000442390.1
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr2_-_180871780 | 0.06 |
ENST00000410053.3
ENST00000295749.6 ENST00000404136.2 |
CWC22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
| chr17_-_66287257 | 0.06 |
ENST00000327268.4
|
SLC16A6
|
solute carrier family 16, member 6 |
| chr9_+_136243264 | 0.06 |
ENST00000371955.1
|
C9orf96
|
chromosome 9 open reading frame 96 |
| chr5_+_134074231 | 0.06 |
ENST00000514518.1
|
CAMLG
|
calcium modulating ligand |
| chr1_-_211848899 | 0.06 |
ENST00000366998.3
ENST00000540251.1 ENST00000366999.4 |
NEK2
|
NIMA-related kinase 2 |
| chr18_+_268148 | 0.06 |
ENST00000581677.1
|
RP11-705O1.8
|
RP11-705O1.8 |
| chr19_-_52552051 | 0.06 |
ENST00000221315.5
|
ZNF432
|
zinc finger protein 432 |
| chr14_-_34931458 | 0.06 |
ENST00000298130.4
|
SPTSSA
|
serine palmitoyltransferase, small subunit A |
| chr20_+_44486246 | 0.06 |
ENST00000255152.2
ENST00000454862.2 |
ZSWIM3
|
zinc finger, SWIM-type containing 3 |
| chr9_+_131217459 | 0.06 |
ENST00000497812.2
ENST00000393533.2 |
ODF2
|
outer dense fiber of sperm tails 2 |
| chr3_+_56591184 | 0.06 |
ENST00000422222.1
ENST00000394672.3 ENST00000326595.7 |
CCDC66
|
coiled-coil domain containing 66 |
| chr1_+_185014496 | 0.06 |
ENST00000367510.3
|
RNF2
|
ring finger protein 2 |
| chr12_-_14849470 | 0.06 |
ENST00000261170.3
|
GUCY2C
|
guanylate cyclase 2C (heat stable enterotoxin receptor) |
| chr19_-_2783255 | 0.06 |
ENST00000589251.1
|
SGTA
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha |
| chr19_-_39826639 | 0.06 |
ENST00000602185.1
ENST00000598034.1 ENST00000601387.1 ENST00000595636.1 ENST00000253054.8 ENST00000594700.1 ENST00000597595.1 |
GMFG
|
glia maturation factor, gamma |
| chr3_+_14989186 | 0.06 |
ENST00000435454.1
ENST00000323373.6 |
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr12_-_96793142 | 0.06 |
ENST00000552262.1
ENST00000551816.1 ENST00000552496.1 |
CDK17
|
cyclin-dependent kinase 17 |
| chr15_-_78526942 | 0.06 |
ENST00000258873.4
|
ACSBG1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr22_+_31518938 | 0.06 |
ENST00000412985.1
ENST00000331075.5 ENST00000412277.2 ENST00000420017.1 ENST00000400294.2 ENST00000405300.1 ENST00000404390.3 |
INPP5J
|
inositol polyphosphate-5-phosphatase J |
| chr7_-_72936531 | 0.06 |
ENST00000339594.4
|
BAZ1B
|
bromodomain adjacent to zinc finger domain, 1B |
| chr14_-_103989033 | 0.06 |
ENST00000553878.1
ENST00000557530.1 |
CKB
|
creatine kinase, brain |
| chr19_-_43702231 | 0.05 |
ENST00000597374.1
ENST00000599371.1 |
PSG4
|
pregnancy specific beta-1-glycoprotein 4 |
| chr2_-_177684007 | 0.05 |
ENST00000451851.1
|
AC092162.1
|
AC092162.1 |
| chr3_-_110612059 | 0.05 |
ENST00000485473.1
|
RP11-553A10.1
|
Uncharacterized protein |
| chr4_+_71859156 | 0.05 |
ENST00000286648.5
ENST00000504730.1 ENST00000504952.1 |
DCK
|
deoxycytidine kinase |
| chr9_-_130966497 | 0.05 |
ENST00000393608.1
ENST00000372948.3 |
CIZ1
|
CDKN1A interacting zinc finger protein 1 |
| chr6_-_31138439 | 0.05 |
ENST00000259915.8
|
POU5F1
|
POU class 5 homeobox 1 |
| chr22_+_20104947 | 0.05 |
ENST00000402752.1
|
RANBP1
|
RAN binding protein 1 |
| chr17_-_40264692 | 0.05 |
ENST00000591220.1
ENST00000251642.3 |
DHX58
|
DEXH (Asp-Glu-X-His) box polypeptide 58 |
| chr17_-_27278445 | 0.05 |
ENST00000268756.3
ENST00000584685.1 |
PHF12
|
PHD finger protein 12 |
| chr6_+_32006042 | 0.05 |
ENST00000418967.2
|
CYP21A2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr3_+_183894737 | 0.05 |
ENST00000432591.1
ENST00000431779.1 |
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit |
| chr19_-_58485895 | 0.05 |
ENST00000314391.3
|
C19orf18
|
chromosome 19 open reading frame 18 |
| chr5_+_140027355 | 0.05 |
ENST00000417647.2
ENST00000507593.1 ENST00000508301.1 |
IK
|
IK cytokine, down-regulator of HLA II |
| chr14_-_21492113 | 0.05 |
ENST00000554094.1
|
NDRG2
|
NDRG family member 2 |
| chr8_+_97597148 | 0.05 |
ENST00000521590.1
|
SDC2
|
syndecan 2 |
| chr5_+_172332220 | 0.05 |
ENST00000518247.1
ENST00000326654.2 |
ERGIC1
|
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
| chr7_-_151433393 | 0.05 |
ENST00000492843.1
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr16_-_68269971 | 0.05 |
ENST00000565858.1
|
ESRP2
|
epithelial splicing regulatory protein 2 |
| chr4_-_76598326 | 0.05 |
ENST00000503660.1
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr3_-_122102065 | 0.05 |
ENST00000479899.1
ENST00000291458.5 ENST00000497726.1 |
CCDC58
|
coiled-coil domain containing 58 |
| chr2_+_69240415 | 0.05 |
ENST00000409829.3
|
ANTXR1
|
anthrax toxin receptor 1 |
| chr3_-_167813132 | 0.05 |
ENST00000309027.4
|
GOLIM4
|
golgi integral membrane protein 4 |
| chr1_-_150693305 | 0.05 |
ENST00000368987.1
|
HORMAD1
|
HORMA domain containing 1 |
| chr7_-_151433342 | 0.05 |
ENST00000433631.2
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr10_+_86088381 | 0.05 |
ENST00000224756.8
ENST00000372088.2 |
CCSER2
|
coiled-coil serine-rich protein 2 |
| chr10_-_31320860 | 0.05 |
ENST00000436087.2
ENST00000442986.1 ENST00000413025.1 ENST00000452305.1 |
ZNF438
|
zinc finger protein 438 |
| chr21_-_27107344 | 0.05 |
ENST00000457143.2
|
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr8_+_96037255 | 0.05 |
ENST00000286687.4
|
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr3_+_13978873 | 0.05 |
ENST00000530586.1
|
TPRXL
|
tetra-peptide repeat homeobox-like |
| chr2_-_61244550 | 0.05 |
ENST00000421319.1
|
PUS10
|
pseudouridylate synthase 10 |
| chr10_+_26986582 | 0.05 |
ENST00000376215.5
ENST00000376203.5 |
PDSS1
|
prenyl (decaprenyl) diphosphate synthase, subunit 1 |
| chr7_-_72936608 | 0.05 |
ENST00000404251.1
|
BAZ1B
|
bromodomain adjacent to zinc finger domain, 1B |
| chr12_-_53729525 | 0.05 |
ENST00000303846.3
|
SP7
|
Sp7 transcription factor |
| chr2_+_234104079 | 0.05 |
ENST00000417661.1
|
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa |
| chr11_+_9482551 | 0.05 |
ENST00000438144.2
ENST00000526657.1 ENST00000299606.2 ENST00000534265.1 ENST00000412390.2 |
ZNF143
|
zinc finger protein 143 |
| chr4_+_103790462 | 0.05 |
ENST00000503643.1
|
CISD2
|
CDGSH iron sulfur domain 2 |
| chr19_+_58193337 | 0.05 |
ENST00000601064.1
ENST00000282296.5 ENST00000356715.4 |
ZNF551
|
zinc finger protein 551 |
| chr15_+_33010175 | 0.05 |
ENST00000300177.4
ENST00000560677.1 ENST00000560830.1 |
GREM1
|
gremlin 1, DAN family BMP antagonist |
| chr17_-_66287310 | 0.05 |
ENST00000582867.1
|
SLC16A6
|
solute carrier family 16, member 6 |
| chr7_-_130597935 | 0.05 |
ENST00000447307.1
ENST00000418546.1 |
MIR29B1
|
microRNA 29a |
| chr3_-_24536453 | 0.05 |
ENST00000453729.2
ENST00000413780.1 |
THRB
|
thyroid hormone receptor, beta |
| chr11_+_57365150 | 0.05 |
ENST00000457869.1
ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr2_+_192543694 | 0.05 |
ENST00000435931.1
|
NABP1
|
nucleic acid binding protein 1 |
| chr11_-_3013482 | 0.05 |
ENST00000529361.1
ENST00000528968.1 ENST00000534372.1 ENST00000531291.1 ENST00000526842.1 |
NAP1L4
|
nucleosome assembly protein 1-like 4 |
| chr17_+_48046671 | 0.05 |
ENST00000505318.2
|
DLX4
|
distal-less homeobox 4 |
| chr11_+_73498973 | 0.05 |
ENST00000537007.1
|
MRPL48
|
mitochondrial ribosomal protein L48 |
| chr10_-_31320840 | 0.05 |
ENST00000375311.1
|
ZNF438
|
zinc finger protein 438 |
| chr14_-_55369525 | 0.05 |
ENST00000543643.2
ENST00000536224.2 ENST00000395514.1 ENST00000491895.2 |
GCH1
|
GTP cyclohydrolase 1 |
| chr22_+_20105012 | 0.05 |
ENST00000331821.3
ENST00000411892.1 |
RANBP1
|
RAN binding protein 1 |
| chr3_+_13978886 | 0.05 |
ENST00000524375.1
ENST00000326972.8 |
TPRXL
|
tetra-peptide repeat homeobox-like |
| chr13_+_49822041 | 0.05 |
ENST00000538056.1
ENST00000251108.6 ENST00000444959.1 ENST00000429346.1 |
CDADC1
|
cytidine and dCMP deaminase domain containing 1 |
| chr9_+_135937365 | 0.05 |
ENST00000372080.4
ENST00000351304.7 |
CEL
|
carboxyl ester lipase |
| chr21_-_35340759 | 0.05 |
ENST00000607953.1
|
AP000569.9
|
AP000569.9 |
| chr1_+_46769303 | 0.05 |
ENST00000311672.5
|
UQCRH
|
ubiquinol-cytochrome c reductase hinge protein |
| chr17_+_38337491 | 0.05 |
ENST00000538981.1
|
RAPGEFL1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr3_-_167452262 | 0.05 |
ENST00000487947.2
|
PDCD10
|
programmed cell death 10 |
| chr1_-_26233423 | 0.05 |
ENST00000357865.2
|
STMN1
|
stathmin 1 |
| chr15_+_59730348 | 0.05 |
ENST00000288228.5
ENST00000559628.1 ENST00000557914.1 ENST00000560474.1 |
FAM81A
|
family with sequence similarity 81, member A |
| chr8_-_124286735 | 0.05 |
ENST00000395571.3
|
ZHX1
|
zinc fingers and homeoboxes 1 |
| chr7_-_8301682 | 0.05 |
ENST00000396675.3
ENST00000430867.1 |
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr5_-_145483932 | 0.05 |
ENST00000311450.4
|
PLAC8L1
|
PLAC8-like 1 |
| chr1_+_68150744 | 0.05 |
ENST00000370986.4
ENST00000370985.3 |
GADD45A
|
growth arrest and DNA-damage-inducible, alpha |
| chr5_+_218356 | 0.05 |
ENST00000264932.6
ENST00000504309.1 ENST00000510361.1 |
SDHA
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp) |
| chr4_+_146560245 | 0.05 |
ENST00000541599.1
|
MMAA
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr3_+_154797781 | 0.04 |
ENST00000382989.3
|
MME
|
membrane metallo-endopeptidase |
| chr22_+_20105259 | 0.04 |
ENST00000416427.1
ENST00000421656.1 ENST00000423859.1 ENST00000418705.2 |
RANBP1
|
RAN binding protein 1 |
| chr7_-_105925558 | 0.04 |
ENST00000222553.3
|
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chr1_+_154947126 | 0.04 |
ENST00000368439.1
|
CKS1B
|
CDC28 protein kinase regulatory subunit 1B |
| chr16_-_3137080 | 0.04 |
ENST00000574387.1
ENST00000571404.1 |
RP11-473M20.9
|
RP11-473M20.9 |
| chr8_+_26149007 | 0.04 |
ENST00000380737.3
ENST00000524169.1 |
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha |
| chr4_-_100867864 | 0.04 |
ENST00000442697.2
|
DNAJB14
|
DnaJ (Hsp40) homolog, subfamily B, member 14 |
| chr8_-_94928861 | 0.04 |
ENST00000607097.1
|
MIR378D2
|
microRNA 378d-2 |
| chr6_+_155537771 | 0.04 |
ENST00000275246.7
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr5_+_159656437 | 0.04 |
ENST00000402432.3
|
FABP6
|
fatty acid binding protein 6, ileal |
| chr9_+_74764278 | 0.04 |
ENST00000238018.4
ENST00000376989.3 |
GDA
|
guanine deaminase |
| chr17_+_66244071 | 0.04 |
ENST00000580548.1
ENST00000580753.1 ENST00000392720.2 ENST00000359783.4 ENST00000584837.1 ENST00000579724.1 ENST00000584494.1 ENST00000580837.1 |
AMZ2
|
archaelysin family metallopeptidase 2 |
| chr4_-_147442817 | 0.04 |
ENST00000507030.1
|
SLC10A7
|
solute carrier family 10, member 7 |
| chr14_-_35183755 | 0.04 |
ENST00000555765.1
|
CFL2
|
cofilin 2 (muscle) |
| chr9_+_74526384 | 0.04 |
ENST00000334731.2
ENST00000377031.3 |
C9orf85
|
chromosome 9 open reading frame 85 |
| chr4_-_668108 | 0.04 |
ENST00000304312.4
|
ATP5I
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit E |
| chr2_+_112895939 | 0.04 |
ENST00000331203.2
ENST00000409903.1 ENST00000409667.3 ENST00000409450.3 |
FBLN7
|
fibulin 7 |
| chr2_+_162016827 | 0.04 |
ENST00000429217.1
ENST00000406287.1 ENST00000402568.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr3_-_167452298 | 0.04 |
ENST00000475915.2
ENST00000462725.2 ENST00000461494.1 |
PDCD10
|
programmed cell death 10 |
| chr12_-_120663792 | 0.04 |
ENST00000546532.1
ENST00000548912.1 |
PXN
|
paxillin |
| chr8_-_124286495 | 0.04 |
ENST00000297857.2
|
ZHX1
|
zinc fingers and homeoboxes 1 |
| chr16_+_67840986 | 0.04 |
ENST00000561639.1
ENST00000567852.1 ENST00000565148.1 ENST00000388833.3 ENST00000561654.1 ENST00000431934.2 |
TSNAXIP1
|
translin-associated factor X interacting protein 1 |
| chr1_-_91487770 | 0.04 |
ENST00000337393.5
|
ZNF644
|
zinc finger protein 644 |
| chr7_-_100823496 | 0.04 |
ENST00000455377.1
ENST00000443096.1 ENST00000300303.2 |
NAT16
|
N-acetyltransferase 16 (GCN5-related, putative) |
| chr4_+_170541678 | 0.04 |
ENST00000360642.3
ENST00000512813.1 |
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr7_+_128399002 | 0.04 |
ENST00000493278.1
|
CALU
|
calumenin |
| chr8_-_144413981 | 0.04 |
ENST00000522041.1
|
TOP1MT
|
topoisomerase (DNA) I, mitochondrial |
| chr2_+_69240511 | 0.04 |
ENST00000409349.3
|
ANTXR1
|
anthrax toxin receptor 1 |
| chr7_-_8301768 | 0.04 |
ENST00000265577.7
|
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr13_-_48575376 | 0.04 |
ENST00000434484.1
|
SUCLA2
|
succinate-CoA ligase, ADP-forming, beta subunit |
| chr2_+_198365095 | 0.04 |
ENST00000409468.1
|
HSPE1
|
heat shock 10kDa protein 1 |
| chr12_+_93963590 | 0.04 |
ENST00000340600.2
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chr8_-_131028641 | 0.04 |
ENST00000523509.1
|
FAM49B
|
family with sequence similarity 49, member B |
| chr1_+_212738676 | 0.04 |
ENST00000366981.4
ENST00000366987.2 |
ATF3
|
activating transcription factor 3 |
| chr19_-_4535233 | 0.04 |
ENST00000381848.3
ENST00000588887.1 ENST00000586133.1 |
PLIN5
|
perilipin 5 |
| chr11_-_69867159 | 0.04 |
ENST00000528507.1
|
RP11-626H12.2
|
RP11-626H12.2 |
| chr7_-_86848933 | 0.04 |
ENST00000423734.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr8_+_109455845 | 0.04 |
ENST00000220853.3
|
EMC2
|
ER membrane protein complex subunit 2 |
| chr16_-_74734742 | 0.04 |
ENST00000308807.7
ENST00000573267.1 |
MLKL
|
mixed lineage kinase domain-like |
| chr5_-_111754948 | 0.04 |
ENST00000261486.5
|
EPB41L4A
|
erythrocyte membrane protein band 4.1 like 4A |
| chrX_-_71497148 | 0.04 |
ENST00000316084.6
|
RPS4X
|
ribosomal protein S4, X-linked |
| chr8_-_80942139 | 0.04 |
ENST00000521434.1
ENST00000519120.1 ENST00000520946.1 |
MRPS28
|
mitochondrial ribosomal protein S28 |
| chr11_+_73498898 | 0.04 |
ENST00000535529.1
ENST00000497094.2 ENST00000411840.2 ENST00000535277.1 ENST00000398483.3 ENST00000542303.1 |
MRPL48
|
mitochondrial ribosomal protein L48 |
| chr2_+_33701707 | 0.04 |
ENST00000425210.1
ENST00000444784.1 ENST00000423159.1 |
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr17_-_7082861 | 0.04 |
ENST00000269299.3
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr13_+_21714913 | 0.04 |
ENST00000450573.1
ENST00000467636.1 |
SAP18
|
Sin3A-associated protein, 18kDa |
| chr15_-_52404921 | 0.04 |
ENST00000561198.1
ENST00000260442.3 |
BCL2L10
|
BCL2-like 10 (apoptosis facilitator) |
| chr7_-_105925367 | 0.04 |
ENST00000354289.4
|
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chr10_+_76586348 | 0.04 |
ENST00000372724.1
ENST00000287239.4 ENST00000372714.1 |
KAT6B
|
K(lysine) acetyltransferase 6B |
| chr9_+_117350009 | 0.04 |
ENST00000374050.3
|
ATP6V1G1
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G1 |
| chr2_+_207024306 | 0.04 |
ENST00000236957.5
ENST00000392221.1 ENST00000392222.2 ENST00000445505.1 |
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr2_+_219433588 | 0.04 |
ENST00000295701.5
|
RQCD1
|
RCD1 required for cell differentiation1 homolog (S. pombe) |
| chr14_-_51027838 | 0.04 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr6_-_138833630 | 0.04 |
ENST00000533765.1
|
NHSL1
|
NHS-like 1 |
| chr19_-_5624057 | 0.04 |
ENST00000590262.1
|
SAFB2
|
scaffold attachment factor B2 |
| chr1_-_38218577 | 0.04 |
ENST00000540011.1
|
EPHA10
|
EPH receptor A10 |
| chr1_+_153950202 | 0.04 |
ENST00000608236.1
|
RP11-422P24.11
|
RP11-422P24.11 |
| chr14_-_24729251 | 0.04 |
ENST00000559136.1
|
TGM1
|
transglutaminase 1 |
| chr3_-_45957534 | 0.04 |
ENST00000536047.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr3_-_138312971 | 0.04 |
ENST00000485115.1
ENST00000484888.1 ENST00000468900.1 ENST00000542237.1 ENST00000481834.1 |
CEP70
|
centrosomal protein 70kDa |
Network of associatons between targets according to the STRING database.
First level regulatory network of RARG
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.0 | 0.1 | GO:0060629 | meiotic DNA double-strand break formation(GO:0042138) regulation of homologous chromosome segregation(GO:0060629) |
| 0.0 | 0.3 | GO:2000567 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.0 | 0.1 | GO:1903565 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.0 | 0.1 | GO:0006147 | guanine catabolic process(GO:0006147) |
| 0.0 | 0.0 | GO:0046449 | creatinine metabolic process(GO:0046449) |
| 0.0 | 0.0 | GO:0061083 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.0 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.0 | 0.1 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
| 0.0 | 0.0 | GO:1903019 | negative regulation of glycoprotein biosynthetic process(GO:0010561) negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) negative regulation of glycoprotein metabolic process(GO:1903019) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.1 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.1 | GO:0060032 | notochord regression(GO:0060032) |
| 0.0 | 0.0 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.0 | 0.1 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.0 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.0 | 0.0 | GO:1903280 | negative regulation of calcium:sodium antiporter activity(GO:1903280) |
| 0.0 | 0.0 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.0 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.0 | GO:0045362 | interleukin-1 biosynthetic process(GO:0042222) regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.0 | 0.1 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.1 | GO:0032425 | positive regulation of mismatch repair(GO:0032425) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.0 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 | 0.1 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.0 | 0.0 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.0 | 0.0 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.0 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.0 | GO:1900155 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.0 | 0.0 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.0 | 0.0 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.0 | 0.0 | GO:0035508 | positive regulation of myosin-light-chain-phosphatase activity(GO:0035508) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.1 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.0 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.0 | 0.1 | GO:0042701 | progesterone secretion(GO:0042701) |
| 0.0 | 0.1 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.0 | GO:0060164 | negative regulation of auditory receptor cell differentiation(GO:0045608) negative regulation of mechanoreceptor differentiation(GO:0045632) regulation of timing of neuron differentiation(GO:0060164) negative regulation of inner ear receptor cell differentiation(GO:2000981) |
| 0.0 | 0.1 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.0 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.0 | 0.1 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.1 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.0 | 0.0 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.3 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.0 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.0 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.0 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.0 | GO:0031379 | RNA-directed RNA polymerase complex(GO:0031379) |
| 0.0 | 0.0 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.0 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.0 | 0.3 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0008892 | guanine deaminase activity(GO:0008892) |
| 0.0 | 0.1 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.1 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.0 | 0.0 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.0 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.0 | GO:0004397 | histidine ammonia-lyase activity(GO:0004397) |
| 0.0 | 0.0 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.0 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.0 | GO:0048244 | phytanoyl-CoA dioxygenase activity(GO:0048244) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.0 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.1 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.1 | GO:0005497 | androgen binding(GO:0005497) |