Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
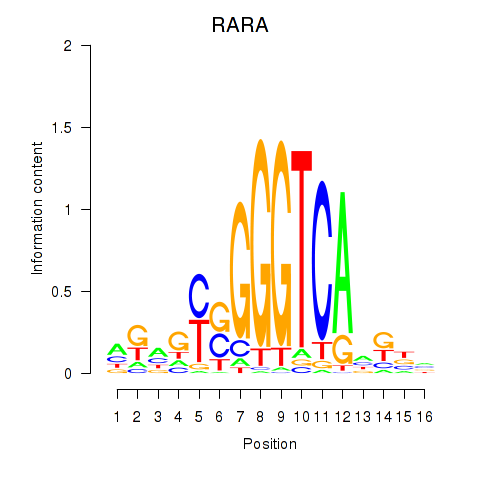
Results for RARA
Z-value: 0.69
Transcription factors associated with RARA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RARA
|
ENSG00000131759.13 | retinoic acid receptor alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RARA | hg19_v2_chr17_+_38497640_38497647 | -0.96 | 3.6e-02 | Click! |
Activity profile of RARA motif
Sorted Z-values of RARA motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_44526786 | 0.39 |
ENST00000245121.5
ENST00000356157.7 |
KATNAL2
|
katanin p60 subunit A-like 2 |
| chr22_+_29664241 | 0.36 |
ENST00000436425.1
ENST00000447973.1 |
EWSR1
|
EWS RNA-binding protein 1 |
| chr8_-_29120604 | 0.26 |
ENST00000521515.1
|
KIF13B
|
kinesin family member 13B |
| chr5_-_31532238 | 0.26 |
ENST00000507438.1
|
DROSHA
|
drosha, ribonuclease type III |
| chr10_-_43633747 | 0.25 |
ENST00000609407.1
|
RP11-351D16.3
|
RP11-351D16.3 |
| chr2_-_45162783 | 0.23 |
ENST00000432125.2
|
RP11-89K21.1
|
RP11-89K21.1 |
| chr12_-_122107549 | 0.22 |
ENST00000355329.3
|
MORN3
|
MORN repeat containing 3 |
| chr11_-_12030681 | 0.22 |
ENST00000529338.1
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr13_-_52026730 | 0.21 |
ENST00000420668.2
|
INTS6
|
integrator complex subunit 6 |
| chr17_-_16557128 | 0.20 |
ENST00000423860.2
ENST00000311331.7 ENST00000583766.1 |
ZNF624
|
zinc finger protein 624 |
| chr16_-_1429010 | 0.20 |
ENST00000513783.1
|
UNKL
|
unkempt family zinc finger-like |
| chr15_+_30375158 | 0.20 |
ENST00000341650.6
ENST00000567927.1 |
GOLGA8J
|
golgin A8 family, member J |
| chr18_+_44526744 | 0.19 |
ENST00000585469.1
|
KATNAL2
|
katanin p60 subunit A-like 2 |
| chr16_+_71879861 | 0.19 |
ENST00000427980.2
ENST00000568581.1 |
ATXN1L
IST1
|
ataxin 1-like increased sodium tolerance 1 homolog (yeast) |
| chr11_-_61196858 | 0.19 |
ENST00000413184.2
|
CPSF7
|
cleavage and polyadenylation specific factor 7, 59kDa |
| chr2_+_242913327 | 0.19 |
ENST00000426962.1
|
AC093642.3
|
AC093642.3 |
| chr6_-_134861089 | 0.19 |
ENST00000606039.1
|
RP11-557H15.4
|
RP11-557H15.4 |
| chr1_-_85156417 | 0.18 |
ENST00000422026.1
|
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr7_+_30068260 | 0.18 |
ENST00000440706.2
|
PLEKHA8
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 8 |
| chr14_-_45603657 | 0.18 |
ENST00000396062.3
|
FKBP3
|
FK506 binding protein 3, 25kDa |
| chr22_-_24641027 | 0.18 |
ENST00000398292.3
ENST00000263112.7 ENST00000418439.2 ENST00000424217.1 ENST00000327365.4 |
GGT5
|
gamma-glutamyltransferase 5 |
| chr14_-_54955376 | 0.18 |
ENST00000553333.1
|
GMFB
|
glia maturation factor, beta |
| chr10_-_112678904 | 0.18 |
ENST00000423273.1
ENST00000436562.1 ENST00000447005.1 ENST00000454061.1 |
BBIP1
|
BBSome interacting protein 1 |
| chr1_-_11918988 | 0.18 |
ENST00000376468.3
|
NPPB
|
natriuretic peptide B |
| chr9_+_139873264 | 0.18 |
ENST00000446677.1
|
PTGDS
|
prostaglandin D2 synthase 21kDa (brain) |
| chr9_+_114393581 | 0.17 |
ENST00000313525.3
|
DNAJC25
|
DnaJ (Hsp40) homolog, subfamily C , member 25 |
| chr7_-_76247617 | 0.17 |
ENST00000441393.1
|
POMZP3
|
POM121 and ZP3 fusion |
| chr4_-_83934078 | 0.17 |
ENST00000505397.1
|
LIN54
|
lin-54 homolog (C. elegans) |
| chr17_-_34122596 | 0.17 |
ENST00000250144.8
|
MMP28
|
matrix metallopeptidase 28 |
| chr12_-_44152551 | 0.16 |
ENST00000416848.2
ENST00000550784.1 ENST00000547156.1 ENST00000549868.1 ENST00000553166.1 ENST00000551923.1 ENST00000431332.3 ENST00000344862.5 |
PUS7L
|
pseudouridylate synthase 7 homolog (S. cerevisiae)-like |
| chr4_-_83933999 | 0.16 |
ENST00000510557.1
|
LIN54
|
lin-54 homolog (C. elegans) |
| chr4_-_186125077 | 0.16 |
ENST00000458385.2
ENST00000514798.1 ENST00000296775.6 |
KIAA1430
|
KIAA1430 |
| chr17_+_33914424 | 0.16 |
ENST00000590432.1
|
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr15_-_52263937 | 0.16 |
ENST00000315141.5
ENST00000299601.5 |
LEO1
|
Leo1, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae) |
| chr22_+_21400229 | 0.16 |
ENST00000342608.4
ENST00000543388.1 ENST00000442047.1 |
AC002472.13
|
Leucine-rich repeat-containing protein LOC400891 |
| chr5_-_157286104 | 0.16 |
ENST00000530742.1
ENST00000523908.1 ENST00000523094.1 ENST00000296951.5 ENST00000411809.2 |
CLINT1
|
clathrin interactor 1 |
| chr19_+_46144884 | 0.16 |
ENST00000593161.1
|
AC006132.1
|
chromosome 19 open reading frame 83 |
| chr2_+_189157498 | 0.16 |
ENST00000359135.3
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr7_+_69064566 | 0.15 |
ENST00000403018.2
|
AUTS2
|
autism susceptibility candidate 2 |
| chr1_+_113933581 | 0.15 |
ENST00000307546.9
ENST00000369615.1 ENST00000369611.4 |
MAGI3
|
membrane associated guanylate kinase, WW and PDZ domain containing 3 |
| chr7_+_106810165 | 0.15 |
ENST00000468401.1
ENST00000497535.1 ENST00000485846.1 |
HBP1
|
HMG-box transcription factor 1 |
| chr1_-_156675368 | 0.15 |
ENST00000368222.3
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr7_-_100239132 | 0.15 |
ENST00000223051.3
ENST00000431692.1 |
TFR2
|
transferrin receptor 2 |
| chr9_-_132597529 | 0.15 |
ENST00000372447.3
|
C9orf78
|
chromosome 9 open reading frame 78 |
| chr14_-_75536182 | 0.14 |
ENST00000555463.1
|
ACYP1
|
acylphosphatase 1, erythrocyte (common) type |
| chr11_-_130184470 | 0.14 |
ENST00000357899.4
ENST00000397753.1 |
ZBTB44
|
zinc finger and BTB domain containing 44 |
| chr7_+_151038850 | 0.14 |
ENST00000355851.4
ENST00000566856.1 ENST00000470229.1 |
NUB1
|
negative regulator of ubiquitin-like proteins 1 |
| chr6_-_41040195 | 0.14 |
ENST00000463088.1
ENST00000469104.1 ENST00000486443.1 |
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chr7_+_151038785 | 0.14 |
ENST00000413040.2
ENST00000568733.1 |
NUB1
|
negative regulator of ubiquitin-like proteins 1 |
| chr8_+_22411931 | 0.14 |
ENST00000523402.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr10_+_115438920 | 0.14 |
ENST00000429617.1
ENST00000369331.4 |
CASP7
|
caspase 7, apoptosis-related cysteine peptidase |
| chr13_+_43597269 | 0.14 |
ENST00000379221.2
|
DNAJC15
|
DnaJ (Hsp40) homolog, subfamily C, member 15 |
| chr6_-_110500826 | 0.14 |
ENST00000265601.3
ENST00000447287.1 ENST00000444391.1 |
WASF1
|
WAS protein family, member 1 |
| chr6_+_96025341 | 0.14 |
ENST00000369293.1
ENST00000358812.4 |
MANEA
|
mannosidase, endo-alpha |
| chr1_-_59165763 | 0.14 |
ENST00000472487.1
|
MYSM1
|
Myb-like, SWIRM and MPN domains 1 |
| chrX_-_37706815 | 0.13 |
ENST00000378578.4
|
DYNLT3
|
dynein, light chain, Tctex-type 3 |
| chr13_+_103451548 | 0.13 |
ENST00000419638.1
|
BIVM
|
basic, immunoglobulin-like variable motif containing |
| chr2_-_208030886 | 0.13 |
ENST00000426163.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr6_+_133562472 | 0.13 |
ENST00000430974.2
ENST00000367895.5 ENST00000355167.3 ENST00000355286.6 |
EYA4
|
eyes absent homolog 4 (Drosophila) |
| chr15_+_33010175 | 0.13 |
ENST00000300177.4
ENST00000560677.1 ENST00000560830.1 |
GREM1
|
gremlin 1, DAN family BMP antagonist |
| chr3_-_101232019 | 0.13 |
ENST00000394095.2
ENST00000394091.1 ENST00000394094.2 ENST00000358203.3 ENST00000348610.3 ENST00000314261.7 |
SENP7
|
SUMO1/sentrin specific peptidase 7 |
| chr22_+_24951436 | 0.13 |
ENST00000215829.3
|
SNRPD3
|
small nuclear ribonucleoprotein D3 polypeptide 18kDa |
| chr7_+_130794878 | 0.13 |
ENST00000416992.2
|
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr22_+_50312379 | 0.13 |
ENST00000407217.3
ENST00000403427.3 |
CRELD2
|
cysteine-rich with EGF-like domains 2 |
| chr11_+_6947720 | 0.13 |
ENST00000414517.2
|
ZNF215
|
zinc finger protein 215 |
| chr6_-_146285221 | 0.13 |
ENST00000367503.3
ENST00000438092.2 ENST00000275233.7 |
SHPRH
|
SNF2 histone linker PHD RING helicase, E3 ubiquitin protein ligase |
| chr2_+_209131059 | 0.13 |
ENST00000422495.1
ENST00000452564.1 |
PIKFYVE
|
phosphoinositide kinase, FYVE finger containing |
| chr5_-_87564620 | 0.12 |
ENST00000506536.1
ENST00000512429.1 ENST00000514135.1 ENST00000296595.6 ENST00000509387.1 |
TMEM161B
|
transmembrane protein 161B |
| chr5_+_131892815 | 0.12 |
ENST00000453394.1
|
RAD50
|
RAD50 homolog (S. cerevisiae) |
| chr8_-_74659068 | 0.12 |
ENST00000523558.1
ENST00000521210.1 ENST00000355780.5 ENST00000524104.1 ENST00000521736.1 ENST00000521447.1 ENST00000517542.1 ENST00000521451.1 ENST00000521419.1 ENST00000518502.1 ENST00000524300.1 |
STAU2
|
staufen double-stranded RNA binding protein 2 |
| chr4_-_10459009 | 0.12 |
ENST00000507515.1
|
ZNF518B
|
zinc finger protein 518B |
| chr9_-_88969339 | 0.12 |
ENST00000375960.2
ENST00000375961.2 |
ZCCHC6
|
zinc finger, CCHC domain containing 6 |
| chr11_-_12030746 | 0.12 |
ENST00000533813.1
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr18_+_21529811 | 0.12 |
ENST00000588004.1
|
LAMA3
|
laminin, alpha 3 |
| chr5_+_1801503 | 0.12 |
ENST00000274137.5
ENST00000469176.1 |
NDUFS6
|
NADH dehydrogenase (ubiquinone) Fe-S protein 6, 13kDa (NADH-coenzyme Q reductase) |
| chr7_+_30067973 | 0.12 |
ENST00000258679.7
ENST00000449726.1 ENST00000396257.2 ENST00000396259.1 |
PLEKHA8
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 8 |
| chr9_-_86432547 | 0.12 |
ENST00000376365.3
ENST00000376371.2 |
GKAP1
|
G kinase anchoring protein 1 |
| chr10_+_43633914 | 0.12 |
ENST00000374466.3
ENST00000374464.1 |
CSGALNACT2
|
chondroitin sulfate N-acetylgalactosaminyltransferase 2 |
| chr9_-_88969303 | 0.12 |
ENST00000277141.6
ENST00000375963.3 |
ZCCHC6
|
zinc finger, CCHC domain containing 6 |
| chr6_-_146285455 | 0.12 |
ENST00000367505.2
|
SHPRH
|
SNF2 histone linker PHD RING helicase, E3 ubiquitin protein ligase |
| chr15_+_76135622 | 0.12 |
ENST00000338677.4
ENST00000267938.4 ENST00000569423.1 |
UBE2Q2
|
ubiquitin-conjugating enzyme E2Q family member 2 |
| chr2_+_189157536 | 0.12 |
ENST00000409580.1
ENST00000409637.3 |
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr10_-_112678692 | 0.12 |
ENST00000605742.1
|
BBIP1
|
BBSome interacting protein 1 |
| chr16_-_14724131 | 0.12 |
ENST00000341484.7
|
PARN
|
poly(A)-specific ribonuclease |
| chr1_+_113933371 | 0.12 |
ENST00000369617.4
|
MAGI3
|
membrane associated guanylate kinase, WW and PDZ domain containing 3 |
| chr1_+_91966656 | 0.12 |
ENST00000428239.1
ENST00000426137.1 |
CDC7
|
cell division cycle 7 |
| chr15_+_82722225 | 0.12 |
ENST00000300515.8
|
GOLGA6L9
|
golgin A6 family-like 9 |
| chr14_-_71107921 | 0.12 |
ENST00000553982.1
ENST00000500016.1 |
CTD-2540L5.5
CTD-2540L5.6
|
CTD-2540L5.5 CTD-2540L5.6 |
| chr19_+_38880695 | 0.11 |
ENST00000587947.1
ENST00000338502.4 |
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr11_-_18610275 | 0.11 |
ENST00000543987.1
|
UEVLD
|
UEV and lactate/malate dehyrogenase domains |
| chr5_-_43313269 | 0.11 |
ENST00000511774.1
|
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr3_-_52486841 | 0.11 |
ENST00000496590.1
|
TNNC1
|
troponin C type 1 (slow) |
| chr6_-_41040268 | 0.11 |
ENST00000373154.2
ENST00000244558.9 ENST00000464633.1 ENST00000424266.2 ENST00000479950.1 ENST00000482515.1 |
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chr6_+_139349817 | 0.11 |
ENST00000367660.3
|
ABRACL
|
ABRA C-terminal like |
| chr14_+_78227105 | 0.11 |
ENST00000439131.2
ENST00000355883.3 ENST00000557011.1 ENST00000556047.1 |
C14orf178
|
chromosome 14 open reading frame 178 |
| chr4_+_25915896 | 0.11 |
ENST00000514384.1
|
SMIM20
|
small integral membrane protein 20 |
| chr1_+_109289279 | 0.11 |
ENST00000370008.3
|
STXBP3
|
syntaxin binding protein 3 |
| chr6_+_134273321 | 0.11 |
ENST00000457715.1
|
TBPL1
|
TBP-like 1 |
| chrX_+_70503526 | 0.11 |
ENST00000413858.1
ENST00000450092.1 |
NONO
|
non-POU domain containing, octamer-binding |
| chr22_-_29138386 | 0.11 |
ENST00000544772.1
|
CHEK2
|
checkpoint kinase 2 |
| chr7_+_130794846 | 0.11 |
ENST00000421797.2
|
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr3_-_47205457 | 0.11 |
ENST00000409792.3
|
SETD2
|
SET domain containing 2 |
| chr10_-_112678976 | 0.11 |
ENST00000448814.1
|
BBIP1
|
BBSome interacting protein 1 |
| chr4_-_159592996 | 0.11 |
ENST00000508457.1
|
C4orf46
|
chromosome 4 open reading frame 46 |
| chr4_+_76439095 | 0.11 |
ENST00000506261.1
|
THAP6
|
THAP domain containing 6 |
| chr20_-_3644046 | 0.11 |
ENST00000290417.2
ENST00000319242.3 |
GFRA4
|
GDNF family receptor alpha 4 |
| chr17_+_2207238 | 0.11 |
ENST00000575840.1
ENST00000576620.1 ENST00000576848.1 ENST00000574987.1 |
SRR
|
serine racemase |
| chr1_+_91966384 | 0.10 |
ENST00000430031.2
ENST00000234626.6 |
CDC7
|
cell division cycle 7 |
| chr5_-_31532160 | 0.10 |
ENST00000511367.2
ENST00000513349.1 |
DROSHA
|
drosha, ribonuclease type III |
| chr12_+_54378849 | 0.10 |
ENST00000515593.1
|
HOXC10
|
homeobox C10 |
| chr3_+_44596679 | 0.10 |
ENST00000426540.1
ENST00000431636.1 ENST00000341840.3 ENST00000273320.3 |
ZKSCAN7
|
zinc finger with KRAB and SCAN domains 7 |
| chr20_+_43538756 | 0.10 |
ENST00000537323.1
ENST00000217073.2 |
PABPC1L
|
poly(A) binding protein, cytoplasmic 1-like |
| chr1_-_31666767 | 0.10 |
ENST00000530145.1
|
NKAIN1
|
Na+/K+ transporting ATPase interacting 1 |
| chr11_-_46142615 | 0.10 |
ENST00000529734.1
ENST00000323180.6 |
PHF21A
|
PHD finger protein 21A |
| chr3_+_186288454 | 0.10 |
ENST00000265028.3
|
DNAJB11
|
DnaJ (Hsp40) homolog, subfamily B, member 11 |
| chr6_+_111303218 | 0.10 |
ENST00000441448.2
|
RPF2
|
ribosome production factor 2 homolog (S. cerevisiae) |
| chr8_-_57233103 | 0.10 |
ENST00000303749.3
ENST00000522671.1 |
SDR16C5
|
short chain dehydrogenase/reductase family 16C, member 5 |
| chr13_+_53029564 | 0.10 |
ENST00000468284.1
ENST00000378034.3 ENST00000258607.5 ENST00000378037.5 |
CKAP2
|
cytoskeleton associated protein 2 |
| chr1_-_45965525 | 0.10 |
ENST00000488405.2
ENST00000490551.3 ENST00000432082.1 |
CCDC163P
|
coiled-coil domain containing 163, pseudogene |
| chr5_+_65440032 | 0.10 |
ENST00000334121.6
|
SREK1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr11_+_33037401 | 0.10 |
ENST00000241051.3
|
DEPDC7
|
DEP domain containing 7 |
| chr7_-_148725544 | 0.10 |
ENST00000413966.1
|
PDIA4
|
protein disulfide isomerase family A, member 4 |
| chr1_+_92545862 | 0.10 |
ENST00000370382.3
ENST00000342818.3 |
BTBD8
|
BTB (POZ) domain containing 8 |
| chr2_+_175352114 | 0.10 |
ENST00000444196.1
ENST00000417038.1 ENST00000606406.1 |
AC010894.3
|
AC010894.3 |
| chr1_+_222988406 | 0.10 |
ENST00000448808.1
ENST00000457636.1 ENST00000439440.1 |
RP11-452F19.3
|
RP11-452F19.3 |
| chr15_-_74374891 | 0.10 |
ENST00000290438.3
|
GOLGA6A
|
golgin A6 family, member A |
| chr2_-_219433014 | 0.10 |
ENST00000418019.1
ENST00000454775.1 ENST00000338465.5 ENST00000415516.1 ENST00000258399.3 |
USP37
|
ubiquitin specific peptidase 37 |
| chr1_-_156647189 | 0.10 |
ENST00000368223.3
|
NES
|
nestin |
| chr2_+_85132749 | 0.10 |
ENST00000233143.4
|
TMSB10
|
thymosin beta 10 |
| chr20_+_43538692 | 0.10 |
ENST00000217074.4
ENST00000255136.3 |
PABPC1L
|
poly(A) binding protein, cytoplasmic 1-like |
| chr19_+_57154508 | 0.10 |
ENST00000598409.1
|
SMIM17
|
small integral membrane protein 17 |
| chr4_+_159593418 | 0.10 |
ENST00000507475.1
ENST00000307738.5 |
ETFDH
|
electron-transferring-flavoprotein dehydrogenase |
| chr12_+_22778116 | 0.10 |
ENST00000538218.1
|
ETNK1
|
ethanolamine kinase 1 |
| chr2_-_40006357 | 0.10 |
ENST00000505747.1
|
THUMPD2
|
THUMP domain containing 2 |
| chr8_-_29120580 | 0.10 |
ENST00000524189.1
|
KIF13B
|
kinesin family member 13B |
| chr15_+_80351977 | 0.10 |
ENST00000559157.1
ENST00000561012.1 ENST00000564367.1 ENST00000558494.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chrX_+_48687283 | 0.10 |
ENST00000338270.1
|
ERAS
|
ES cell expressed Ras |
| chr9_+_114393634 | 0.09 |
ENST00000556107.1
ENST00000374294.3 |
DNAJC25
DNAJC25-GNG10
|
DnaJ (Hsp40) homolog, subfamily C , member 25 DNAJC25-GNG10 readthrough |
| chr1_+_29241027 | 0.09 |
ENST00000373797.1
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
| chr19_+_49927006 | 0.09 |
ENST00000576655.1
|
CTD-3148I10.1
|
golgi-associated, olfactory signaling regulator |
| chr15_+_23255242 | 0.09 |
ENST00000450802.3
|
GOLGA8I
|
golgin A8 family, member I |
| chr1_-_3816779 | 0.09 |
ENST00000361605.3
|
C1orf174
|
chromosome 1 open reading frame 174 |
| chr4_-_184580304 | 0.09 |
ENST00000510968.1
ENST00000512740.1 ENST00000327570.9 |
RWDD4
|
RWD domain containing 4 |
| chr11_-_70963538 | 0.09 |
ENST00000413503.1
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr1_+_219347203 | 0.09 |
ENST00000366927.3
|
LYPLAL1
|
lysophospholipase-like 1 |
| chr19_-_7167989 | 0.09 |
ENST00000600492.1
|
INSR
|
insulin receptor |
| chr9_-_79009048 | 0.09 |
ENST00000490113.1
|
RFK
|
riboflavin kinase |
| chr16_+_1359511 | 0.09 |
ENST00000397514.3
ENST00000397515.2 ENST00000567383.1 ENST00000403747.2 ENST00000566587.1 |
UBE2I
|
ubiquitin-conjugating enzyme E2I |
| chr19_+_32836499 | 0.09 |
ENST00000311921.4
ENST00000544431.1 ENST00000355898.5 |
ZNF507
|
zinc finger protein 507 |
| chr12_+_133613878 | 0.09 |
ENST00000392319.2
ENST00000543758.1 |
ZNF84
|
zinc finger protein 84 |
| chr16_-_14724057 | 0.09 |
ENST00000539279.1
ENST00000420015.2 ENST00000437198.2 |
PARN
|
poly(A)-specific ribonuclease |
| chr15_+_74908147 | 0.09 |
ENST00000568139.1
ENST00000563297.1 ENST00000568488.1 ENST00000352989.5 ENST00000348245.3 |
CLK3
|
CDC-like kinase 3 |
| chr6_+_32006042 | 0.09 |
ENST00000418967.2
|
CYP21A2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr17_-_47308128 | 0.09 |
ENST00000413580.1
ENST00000511066.1 |
PHOSPHO1
|
phosphatase, orphan 1 |
| chr5_-_59783882 | 0.09 |
ENST00000505507.2
ENST00000502484.2 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr9_-_100935043 | 0.09 |
ENST00000343933.5
|
CORO2A
|
coronin, actin binding protein, 2A |
| chr1_-_25558984 | 0.09 |
ENST00000236273.4
|
SYF2
|
SYF2 pre-mRNA-splicing factor |
| chr1_+_222988363 | 0.09 |
ENST00000450784.1
ENST00000426045.1 ENST00000457955.1 ENST00000444858.1 ENST00000435378.1 ENST00000441676.1 |
RP11-452F19.3
|
RP11-452F19.3 |
| chr6_+_132129151 | 0.09 |
ENST00000360971.2
|
ENPP1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1 |
| chr1_+_145726886 | 0.09 |
ENST00000443667.1
|
PDZK1
|
PDZ domain containing 1 |
| chrX_-_37706661 | 0.09 |
ENST00000432389.2
ENST00000378581.3 |
DYNLT3
|
dynein, light chain, Tctex-type 3 |
| chr1_-_33168336 | 0.09 |
ENST00000373484.3
|
SYNC
|
syncoilin, intermediate filament protein |
| chr10_+_1034338 | 0.09 |
ENST00000360803.4
ENST00000538293.1 |
GTPBP4
|
GTP binding protein 4 |
| chr7_-_21985656 | 0.09 |
ENST00000406877.3
|
CDCA7L
|
cell division cycle associated 7-like |
| chr16_-_57219721 | 0.09 |
ENST00000562406.1
ENST00000568671.1 ENST00000567044.1 |
FAM192A
|
family with sequence similarity 192, member A |
| chr17_-_8868991 | 0.09 |
ENST00000447110.1
|
PIK3R5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chr15_-_101817492 | 0.09 |
ENST00000528346.1
ENST00000531964.1 |
VIMP
|
VCP-interacting membrane protein |
| chr5_-_43313574 | 0.09 |
ENST00000325110.6
ENST00000433297.2 |
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr11_+_119019722 | 0.09 |
ENST00000307417.3
|
ABCG4
|
ATP-binding cassette, sub-family G (WHITE), member 4 |
| chr19_-_19739007 | 0.08 |
ENST00000586703.1
ENST00000591042.1 ENST00000407877.3 |
LPAR2
|
lysophosphatidic acid receptor 2 |
| chr4_-_184580353 | 0.08 |
ENST00000326397.5
|
RWDD4
|
RWD domain containing 4 |
| chrX_+_2670066 | 0.08 |
ENST00000381174.5
ENST00000419513.2 ENST00000426774.1 |
XG
|
Xg blood group |
| chr22_+_29664248 | 0.08 |
ENST00000406548.1
ENST00000437155.2 ENST00000415761.1 ENST00000331029.7 |
EWSR1
|
EWS RNA-binding protein 1 |
| chr19_-_12405689 | 0.08 |
ENST00000355684.5
|
ZNF44
|
zinc finger protein 44 |
| chr1_-_16302608 | 0.08 |
ENST00000375743.4
ENST00000375733.2 |
ZBTB17
|
zinc finger and BTB domain containing 17 |
| chr19_-_37958318 | 0.08 |
ENST00000316950.6
ENST00000591710.1 |
ZNF569
|
zinc finger protein 569 |
| chr6_-_57086200 | 0.08 |
ENST00000468148.1
|
RAB23
|
RAB23, member RAS oncogene family |
| chr5_-_137911049 | 0.08 |
ENST00000297185.3
|
HSPA9
|
heat shock 70kDa protein 9 (mortalin) |
| chr2_-_113191096 | 0.08 |
ENST00000496537.1
ENST00000330575.5 ENST00000302558.3 |
RGPD8
|
RANBP2-like and GRIP domain containing 8 |
| chr1_-_180991978 | 0.08 |
ENST00000542060.1
ENST00000258301.5 |
STX6
|
syntaxin 6 |
| chr13_+_53030107 | 0.08 |
ENST00000490903.1
ENST00000480747.1 |
CKAP2
|
cytoskeleton associated protein 2 |
| chr9_-_131038266 | 0.08 |
ENST00000490628.1
ENST00000421699.2 ENST00000450617.1 |
GOLGA2
|
golgin A2 |
| chr1_-_109203685 | 0.08 |
ENST00000402983.1
ENST00000420055.1 |
HENMT1
|
HEN1 methyltransferase homolog 1 (Arabidopsis) |
| chr12_+_54378923 | 0.08 |
ENST00000303460.4
|
HOXC10
|
homeobox C10 |
| chr12_-_110011288 | 0.08 |
ENST00000540016.1
ENST00000266839.5 |
MMAB
|
methylmalonic aciduria (cobalamin deficiency) cblB type |
| chr9_-_79009414 | 0.08 |
ENST00000376736.1
|
RFK
|
riboflavin kinase |
| chr5_+_135468516 | 0.08 |
ENST00000507118.1
ENST00000511116.1 ENST00000545279.1 ENST00000545620.1 |
SMAD5
|
SMAD family member 5 |
| chr1_+_171227069 | 0.08 |
ENST00000354841.4
|
FMO1
|
flavin containing monooxygenase 1 |
| chr17_-_80291818 | 0.08 |
ENST00000269389.3
ENST00000581691.1 |
SECTM1
|
secreted and transmembrane 1 |
| chr1_-_211848899 | 0.08 |
ENST00000366998.3
ENST00000540251.1 ENST00000366999.4 |
NEK2
|
NIMA-related kinase 2 |
| chr9_+_21802542 | 0.08 |
ENST00000380172.4
|
MTAP
|
methylthioadenosine phosphorylase |
| chr10_+_70715884 | 0.08 |
ENST00000354185.4
|
DDX21
|
DEAD (Asp-Glu-Ala-Asp) box helicase 21 |
| chr2_-_74555350 | 0.08 |
ENST00000444570.1
|
SLC4A5
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 5 |
| chr2_+_192542879 | 0.08 |
ENST00000409510.1
|
NABP1
|
nucleic acid binding protein 1 |
| chr9_-_125027079 | 0.08 |
ENST00000417201.3
|
RBM18
|
RNA binding motif protein 18 |
| chr11_-_33795893 | 0.07 |
ENST00000526785.1
ENST00000534136.1 ENST00000265651.3 ENST00000530401.1 ENST00000448981.2 |
FBXO3
|
F-box protein 3 |
| chr16_-_57219926 | 0.07 |
ENST00000566584.1
ENST00000566481.1 ENST00000566077.1 ENST00000564108.1 ENST00000565458.1 ENST00000566681.1 ENST00000567439.1 |
FAM192A
|
family with sequence similarity 192, member A |
| chr22_+_50312316 | 0.07 |
ENST00000328268.4
|
CRELD2
|
cysteine-rich with EGF-like domains 2 |
| chr6_-_74233480 | 0.07 |
ENST00000455918.1
|
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr19_+_35168567 | 0.07 |
ENST00000457781.2
ENST00000505163.1 ENST00000505242.1 ENST00000423823.2 ENST00000507959.1 ENST00000446502.2 |
ZNF302
|
zinc finger protein 302 |
Network of associatons between targets according to the STRING database.
First level regulatory network of RARA
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.1 | 0.2 | GO:0042727 | flavin-containing compound biosynthetic process(GO:0042727) |
| 0.1 | 0.2 | GO:0072428 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.1 | 0.4 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.4 | GO:1990262 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.0 | 0.1 | GO:1904562 | phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.1 | GO:0097198 | cell migration involved in vasculogenesis(GO:0035441) histone H3-K36 trimethylation(GO:0097198) |
| 0.0 | 0.1 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.1 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.1 | GO:1900155 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.0 | 0.2 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 0.0 | 0.1 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.2 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.1 | GO:0097354 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 0.2 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.0 | 0.4 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.2 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 0.1 | GO:0006738 | nicotinamide riboside catabolic process(GO:0006738) nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) pyridine nucleoside catabolic process(GO:0070638) |
| 0.0 | 0.1 | GO:0097460 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.0 | 0.1 | GO:1902957 | negative regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902957) |
| 0.0 | 0.1 | GO:0072684 | mitochondrial tRNA 3'-trailer cleavage, endonucleolytic(GO:0072684) |
| 0.0 | 0.4 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.1 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.1 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.0 | 0.2 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.2 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.1 | GO:0015993 | molecular hydrogen transport(GO:0015993) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.1 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.0 | 0.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.1 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.0 | GO:0045720 | negative regulation of integrin biosynthetic process(GO:0045720) |
| 0.0 | 0.0 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.1 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.1 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.0 | 0.1 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.2 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.2 | GO:0008212 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.0 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.2 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.0 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.0 | GO:0070055 | mRNA splicing via endonucleolytic cleavage and ligation involved in unfolded protein response(GO:0030969) mRNA splicing, via endonucleolytic cleavage and ligation(GO:0070054) mRNA endonucleolytic cleavage involved in unfolded protein response(GO:0070055) |
| 0.0 | 0.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.1 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 0.2 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.0 | 0.1 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.0 | GO:0035669 | TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.0 | 0.1 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.0 | 0.1 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 0.0 | 0.2 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 0.2 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.0 | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.1 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.0 | 0.1 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.0 | 0.1 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0070877 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.0 | 0.4 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.1 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.1 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.0 | 0.0 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0005713 | recombination nodule(GO:0005713) |
| 0.0 | 0.1 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.0 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.1 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 0.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.0 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.1 | 0.2 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.1 | 0.3 | GO:0017089 | glycolipid transporter activity(GO:0017089) ceramide binding(GO:0097001) |
| 0.1 | 0.4 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.1 | 0.6 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.2 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.0 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.1 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.0 | 0.1 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.1 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.0 | 0.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.1 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.1 | GO:0000285 | 1-phosphatidylinositol-3-phosphate 5-kinase activity(GO:0000285) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0070363 | mitochondrial light strand promoter sense binding(GO:0070363) |
| 0.0 | 0.1 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.0 | 0.1 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.2 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.1 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.0 | 0.1 | GO:0035529 | phosphodiesterase I activity(GO:0004528) NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.0 | 0.1 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.0 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.1 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) leukemia inhibitory factor receptor activity(GO:0004923) interleukin-6 binding(GO:0019981) |
| 0.0 | 0.1 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.1 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.0 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.0 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.0 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.1 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.0 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.1 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.0 | 0.1 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.4 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |