Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for PTF1A
Z-value: 0.50
Transcription factors associated with PTF1A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PTF1A
|
ENSG00000168267.5 | pancreas associated transcription factor 1a |
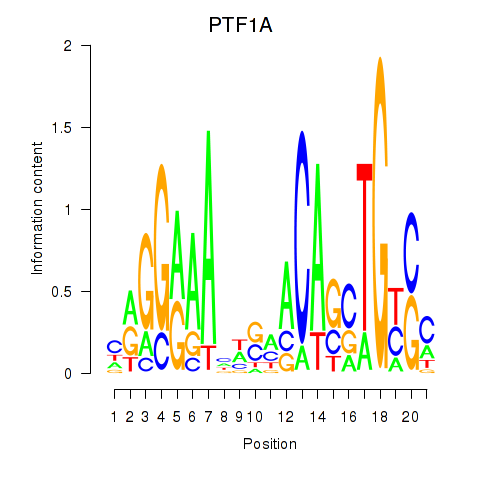
Activity profile of PTF1A motif
Sorted Z-values of PTF1A motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_124739964 | 0.21 |
ENST00000406217.2
|
PSTK
|
phosphoseryl-tRNA kinase |
| chr12_+_124392789 | 0.20 |
ENST00000540041.1
|
DNAH10
|
dynein, axonemal, heavy chain 10 |
| chr4_-_14889791 | 0.18 |
ENST00000509654.1
ENST00000515031.1 ENST00000505089.2 |
LINC00504
|
long intergenic non-protein coding RNA 504 |
| chr2_-_95831158 | 0.17 |
ENST00000447814.1
|
ZNF514
|
zinc finger protein 514 |
| chr3_-_48659193 | 0.17 |
ENST00000330862.3
|
TMEM89
|
transmembrane protein 89 |
| chr16_-_2004683 | 0.17 |
ENST00000268661.7
|
RPL3L
|
ribosomal protein L3-like |
| chr10_-_10504285 | 0.17 |
ENST00000602311.1
|
RP11-271F18.4
|
RP11-271F18.4 |
| chr15_+_84841242 | 0.16 |
ENST00000558195.1
|
RP11-182J1.16
|
ubiquitin-conjugating enzyme E2Q family member 2-like |
| chr16_+_89696692 | 0.15 |
ENST00000261615.4
|
DPEP1
|
dipeptidase 1 (renal) |
| chr3_-_52869205 | 0.15 |
ENST00000446157.2
|
MUSTN1
|
musculoskeletal, embryonic nuclear protein 1 |
| chr19_+_17326521 | 0.15 |
ENST00000593597.1
|
USE1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr8_-_38126635 | 0.14 |
ENST00000529359.1
|
PPAPDC1B
|
phosphatidic acid phosphatase type 2 domain containing 1B |
| chr12_+_2912363 | 0.14 |
ENST00000544366.1
|
FKBP4
|
FK506 binding protein 4, 59kDa |
| chr13_-_31736132 | 0.14 |
ENST00000429785.2
|
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chr3_-_52868931 | 0.13 |
ENST00000486659.1
|
MUSTN1
|
musculoskeletal, embryonic nuclear protein 1 |
| chr21_+_44073860 | 0.13 |
ENST00000335512.4
ENST00000539837.1 ENST00000291539.6 ENST00000380328.2 ENST00000398232.3 ENST00000398234.3 ENST00000398236.3 ENST00000328862.6 ENST00000335440.6 ENST00000398225.3 ENST00000398229.3 ENST00000398227.3 |
PDE9A
|
phosphodiesterase 9A |
| chr11_+_193065 | 0.13 |
ENST00000342878.2
|
SCGB1C1
|
secretoglobin, family 1C, member 1 |
| chr12_+_75784850 | 0.13 |
ENST00000550916.1
ENST00000435775.1 ENST00000378689.2 ENST00000378692.3 ENST00000320460.4 ENST00000547164.1 |
GLIPR1L2
|
GLI pathogenesis-related 1 like 2 |
| chr15_-_89764929 | 0.13 |
ENST00000268125.5
|
RLBP1
|
retinaldehyde binding protein 1 |
| chr15_+_41099919 | 0.12 |
ENST00000561617.1
|
ZFYVE19
|
zinc finger, FYVE domain containing 19 |
| chr1_-_231114542 | 0.12 |
ENST00000522821.1
ENST00000366661.4 ENST00000366662.4 ENST00000414259.1 ENST00000522399.1 |
TTC13
|
tetratricopeptide repeat domain 13 |
| chr1_+_231114795 | 0.12 |
ENST00000310256.2
ENST00000366658.2 ENST00000450711.1 ENST00000435927.1 |
ARV1
|
ARV1 homolog (S. cerevisiae) |
| chr11_-_96123022 | 0.12 |
ENST00000542662.1
|
CCDC82
|
coiled-coil domain containing 82 |
| chr5_-_39425222 | 0.11 |
ENST00000320816.6
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr9_+_107266455 | 0.11 |
ENST00000334726.2
|
OR13F1
|
olfactory receptor, family 13, subfamily F, member 1 |
| chr12_+_113344811 | 0.11 |
ENST00000551241.1
ENST00000553185.1 ENST00000550689.1 |
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr10_+_124739911 | 0.11 |
ENST00000405485.1
|
PSTK
|
phosphoseryl-tRNA kinase |
| chr5_-_162887071 | 0.10 |
ENST00000302764.4
|
NUDCD2
|
NudC domain containing 2 |
| chr1_+_2005126 | 0.10 |
ENST00000495347.1
|
PRKCZ
|
protein kinase C, zeta |
| chr12_+_12966250 | 0.10 |
ENST00000352940.4
ENST00000358007.3 ENST00000544400.1 |
DDX47
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 47 |
| chr19_+_11670245 | 0.10 |
ENST00000585493.1
|
ZNF627
|
zinc finger protein 627 |
| chr1_+_114447763 | 0.10 |
ENST00000369563.3
|
DCLRE1B
|
DNA cross-link repair 1B |
| chr3_-_12883026 | 0.10 |
ENST00000396953.2
ENST00000457131.1 ENST00000435983.1 ENST00000273223.6 ENST00000396957.1 ENST00000429711.2 |
RPL32
|
ribosomal protein L32 |
| chr16_+_71392616 | 0.10 |
ENST00000349553.5
ENST00000302628.4 ENST00000562305.1 |
CALB2
|
calbindin 2 |
| chr14_-_35099377 | 0.10 |
ENST00000362031.4
|
SNX6
|
sorting nexin 6 |
| chr21_+_44073916 | 0.09 |
ENST00000349112.3
ENST00000398224.3 |
PDE9A
|
phosphodiesterase 9A |
| chr10_+_43633914 | 0.09 |
ENST00000374466.3
ENST00000374464.1 |
CSGALNACT2
|
chondroitin sulfate N-acetylgalactosaminyltransferase 2 |
| chr6_-_2962331 | 0.09 |
ENST00000380524.1
|
SERPINB6
|
serpin peptidase inhibitor, clade B (ovalbumin), member 6 |
| chr1_-_114447683 | 0.09 |
ENST00000256658.4
ENST00000369564.1 |
AP4B1
|
adaptor-related protein complex 4, beta 1 subunit |
| chr1_-_114447620 | 0.09 |
ENST00000369569.1
ENST00000432415.1 ENST00000369571.2 |
AP4B1
|
adaptor-related protein complex 4, beta 1 subunit |
| chr19_+_42259329 | 0.09 |
ENST00000199764.6
|
CEACAM6
|
carcinoembryonic antigen-related cell adhesion molecule 6 (non-specific cross reacting antigen) |
| chr7_-_93204033 | 0.09 |
ENST00000359558.2
ENST00000360249.4 ENST00000426151.1 |
CALCR
|
calcitonin receptor |
| chr14_+_67656110 | 0.09 |
ENST00000524532.1
ENST00000530728.1 |
FAM71D
|
family with sequence similarity 71, member D |
| chr14_-_35099315 | 0.09 |
ENST00000396526.3
ENST00000396534.3 ENST00000355110.5 ENST00000557265.1 |
SNX6
|
sorting nexin 6 |
| chr13_-_31736027 | 0.09 |
ENST00000380406.5
ENST00000320027.5 ENST00000380405.4 |
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chr12_-_57644952 | 0.08 |
ENST00000554578.1
ENST00000546246.2 ENST00000553489.1 ENST00000332782.2 |
STAC3
|
SH3 and cysteine rich domain 3 |
| chr5_-_39425290 | 0.08 |
ENST00000545653.1
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr3_-_88108192 | 0.08 |
ENST00000309534.6
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr9_+_4985228 | 0.08 |
ENST00000381652.3
|
JAK2
|
Janus kinase 2 |
| chr12_+_113344582 | 0.08 |
ENST00000202917.5
ENST00000445409.2 ENST00000452357.2 |
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr6_-_134499037 | 0.08 |
ENST00000528577.1
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr11_+_96123158 | 0.08 |
ENST00000332349.4
ENST00000458427.1 |
JRKL
|
jerky homolog-like (mouse) |
| chr12_+_53689309 | 0.08 |
ENST00000351500.3
ENST00000550846.1 ENST00000334478.4 ENST00000549759.1 |
PFDN5
|
prefoldin subunit 5 |
| chr1_+_178694408 | 0.08 |
ENST00000324778.5
|
RALGPS2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr1_+_151682909 | 0.08 |
ENST00000326413.3
ENST00000442233.2 |
RIIAD1
AL589765.1
|
regulatory subunit of type II PKA R-subunit (RIIa) domain containing 1 Uncharacterized protein; cDNA FLJ36032 fis, clone TESTI2017069 |
| chr10_-_5855350 | 0.07 |
ENST00000456041.1
ENST00000380181.3 ENST00000418688.1 ENST00000380132.4 ENST00000609712.1 ENST00000380191.4 |
GDI2
|
GDP dissociation inhibitor 2 |
| chrX_-_44402231 | 0.07 |
ENST00000378045.4
|
FUNDC1
|
FUN14 domain containing 1 |
| chr5_-_118324200 | 0.07 |
ENST00000515439.3
ENST00000510708.1 |
DTWD2
|
DTW domain containing 2 |
| chr15_+_41099788 | 0.07 |
ENST00000299173.10
ENST00000566407.1 |
ZFYVE19
|
zinc finger, FYVE domain containing 19 |
| chr4_-_83719983 | 0.07 |
ENST00000319540.4
|
SCD5
|
stearoyl-CoA desaturase 5 |
| chr14_-_77737543 | 0.07 |
ENST00000298352.4
|
NGB
|
neuroglobin |
| chr7_-_152373216 | 0.07 |
ENST00000359321.1
|
XRCC2
|
X-ray repair complementing defective repair in Chinese hamster cells 2 |
| chrX_-_13956497 | 0.07 |
ENST00000398361.3
|
GPM6B
|
glycoprotein M6B |
| chr15_+_51633826 | 0.07 |
ENST00000335449.6
|
GLDN
|
gliomedin |
| chr6_-_24877490 | 0.07 |
ENST00000540914.1
ENST00000378023.4 |
FAM65B
|
family with sequence similarity 65, member B |
| chr20_-_44937124 | 0.06 |
ENST00000537909.1
|
CDH22
|
cadherin 22, type 2 |
| chr10_-_28571015 | 0.06 |
ENST00000375719.3
ENST00000375732.1 |
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr13_-_111522162 | 0.06 |
ENST00000538077.1
|
LINC00346
|
long intergenic non-protein coding RNA 346 |
| chrX_+_155110956 | 0.06 |
ENST00000286448.6
ENST00000262640.6 ENST00000460621.1 |
VAMP7
|
vesicle-associated membrane protein 7 |
| chr20_+_44441304 | 0.06 |
ENST00000352551.5
|
UBE2C
|
ubiquitin-conjugating enzyme E2C |
| chr3_+_167453493 | 0.06 |
ENST00000295777.5
ENST00000472747.2 |
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr10_-_101841588 | 0.06 |
ENST00000370418.3
|
CPN1
|
carboxypeptidase N, polypeptide 1 |
| chr4_+_20702059 | 0.06 |
ENST00000444671.2
ENST00000510700.1 ENST00000506745.1 ENST00000514663.1 ENST00000509469.1 ENST00000515339.1 ENST00000513861.1 ENST00000502374.1 ENST00000538990.1 ENST00000511160.1 ENST00000504630.1 ENST00000513590.1 ENST00000514292.1 ENST00000502938.1 ENST00000509625.1 ENST00000505160.1 ENST00000507634.1 ENST00000513459.1 ENST00000511089.1 |
PACRGL
|
PARK2 co-regulated-like |
| chr14_-_101351184 | 0.06 |
ENST00000534062.1
|
RTL1
|
retrotransposon-like 1 |
| chr2_+_173600565 | 0.06 |
ENST00000397081.3
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr16_-_85146040 | 0.06 |
ENST00000539556.1
|
FAM92B
|
family with sequence similarity 92, member B |
| chr20_-_48732472 | 0.06 |
ENST00000340309.3
ENST00000415862.2 ENST00000371677.3 ENST00000420027.2 |
UBE2V1
|
ubiquitin-conjugating enzyme E2 variant 1 |
| chr11_-_1643368 | 0.06 |
ENST00000399682.1
|
KRTAP5-4
|
keratin associated protein 5-4 |
| chr14_-_58893832 | 0.06 |
ENST00000556007.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr2_-_136288113 | 0.06 |
ENST00000401392.1
|
ZRANB3
|
zinc finger, RAN-binding domain containing 3 |
| chr2_+_202122826 | 0.06 |
ENST00000413726.1
|
CASP8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr7_-_72993033 | 0.06 |
ENST00000305632.5
|
TBL2
|
transducin (beta)-like 2 |
| chr11_+_8704748 | 0.06 |
ENST00000526562.1
ENST00000525981.1 |
RPL27A
|
ribosomal protein L27a |
| chr5_-_39425068 | 0.05 |
ENST00000515700.1
ENST00000339788.6 |
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr14_-_50154921 | 0.05 |
ENST00000553805.2
ENST00000554396.1 ENST00000216367.5 ENST00000539565.2 |
POLE2
|
polymerase (DNA directed), epsilon 2, accessory subunit |
| chr16_-_46723066 | 0.05 |
ENST00000299138.7
|
VPS35
|
vacuolar protein sorting 35 homolog (S. cerevisiae) |
| chr4_+_183370146 | 0.05 |
ENST00000510504.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chrX_+_154299690 | 0.05 |
ENST00000340647.4
ENST00000330045.7 |
BRCC3
|
BRCA1/BRCA2-containing complex, subunit 3 |
| chr10_+_43572475 | 0.05 |
ENST00000355710.3
ENST00000498820.1 ENST00000340058.5 |
RET
|
ret proto-oncogene |
| chr1_+_100503643 | 0.05 |
ENST00000370152.3
|
HIAT1
|
hippocampus abundant transcript 1 |
| chr2_+_173600514 | 0.05 |
ENST00000264111.6
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr1_+_40204538 | 0.05 |
ENST00000324379.5
ENST00000356511.2 ENST00000497370.1 ENST00000470213.1 ENST00000372835.5 ENST00000372830.1 |
PPIE
|
peptidylprolyl isomerase E (cyclophilin E) |
| chr12_-_48398104 | 0.05 |
ENST00000337299.6
ENST00000380518.3 |
COL2A1
|
collagen, type II, alpha 1 |
| chr4_-_82393009 | 0.05 |
ENST00000436139.2
|
RASGEF1B
|
RasGEF domain family, member 1B |
| chr5_-_75013193 | 0.05 |
ENST00000514838.2
ENST00000506164.1 ENST00000502826.1 ENST00000503835.1 ENST00000428202.2 ENST00000380475.2 |
POC5
|
POC5 centriolar protein |
| chr7_+_7606497 | 0.05 |
ENST00000340080.4
ENST00000405785.1 ENST00000433635.1 |
MIOS
|
missing oocyte, meiosis regulator, homolog (Drosophila) |
| chr1_-_9129631 | 0.05 |
ENST00000377414.3
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr1_+_12185949 | 0.04 |
ENST00000413146.2
|
TNFRSF8
|
tumor necrosis factor receptor superfamily, member 8 |
| chr2_+_202122703 | 0.04 |
ENST00000447616.1
ENST00000358485.4 |
CASP8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr11_+_7597639 | 0.04 |
ENST00000533792.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr20_+_42544782 | 0.04 |
ENST00000423191.2
ENST00000372999.1 |
TOX2
|
TOX high mobility group box family member 2 |
| chrX_+_154299753 | 0.04 |
ENST00000369459.2
ENST00000369462.1 ENST00000411985.1 ENST00000399042.1 |
BRCC3
|
BRCA1/BRCA2-containing complex, subunit 3 |
| chr10_-_74856608 | 0.04 |
ENST00000307116.2
ENST00000373008.2 ENST00000412021.2 ENST00000394890.2 ENST00000263556.3 ENST00000440381.1 |
P4HA1
|
prolyl 4-hydroxylase, alpha polypeptide I |
| chr2_-_196933536 | 0.04 |
ENST00000312428.6
ENST00000410072.1 |
DNAH7
|
dynein, axonemal, heavy chain 7 |
| chr1_-_45956868 | 0.04 |
ENST00000451835.2
|
TESK2
|
testis-specific kinase 2 |
| chr1_+_150480551 | 0.04 |
ENST00000369049.4
ENST00000369047.4 |
ECM1
|
extracellular matrix protein 1 |
| chr14_-_38028689 | 0.04 |
ENST00000553425.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr21_+_39668831 | 0.04 |
ENST00000419868.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr1_+_178694362 | 0.04 |
ENST00000367634.2
|
RALGPS2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr3_+_35681081 | 0.04 |
ENST00000428373.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr12_-_3862347 | 0.04 |
ENST00000444507.1
|
EFCAB4B
|
EF-hand calcium binding domain 4B |
| chr11_+_64058758 | 0.04 |
ENST00000538767.1
|
KCNK4
|
potassium channel, subfamily K, member 4 |
| chr3_+_130648842 | 0.04 |
ENST00000508297.1
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr3_+_101546827 | 0.04 |
ENST00000461724.1
ENST00000483180.1 ENST00000394054.2 |
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr17_-_36499693 | 0.04 |
ENST00000342292.4
|
GPR179
|
G protein-coupled receptor 179 |
| chr6_-_127780510 | 0.04 |
ENST00000487331.2
ENST00000483725.3 |
KIAA0408
|
KIAA0408 |
| chr18_+_33877654 | 0.04 |
ENST00000257209.4
ENST00000445677.1 ENST00000590592.1 ENST00000359247.4 |
FHOD3
|
formin homology 2 domain containing 3 |
| chr19_+_19779686 | 0.03 |
ENST00000415784.2
|
ZNF101
|
zinc finger protein 101 |
| chr10_-_43633747 | 0.03 |
ENST00000609407.1
|
RP11-351D16.3
|
RP11-351D16.3 |
| chrX_-_107019181 | 0.03 |
ENST00000315660.4
ENST00000372384.2 ENST00000502650.1 ENST00000506724.1 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr3_-_195938256 | 0.03 |
ENST00000296326.3
|
ZDHHC19
|
zinc finger, DHHC-type containing 19 |
| chr2_-_86422523 | 0.03 |
ENST00000442664.2
ENST00000409051.2 ENST00000449247.2 |
IMMT
|
inner membrane protein, mitochondrial |
| chr2_+_97426631 | 0.03 |
ENST00000377075.2
|
CNNM4
|
cyclin M4 |
| chr1_+_150480576 | 0.03 |
ENST00000346569.6
|
ECM1
|
extracellular matrix protein 1 |
| chr11_-_1606513 | 0.03 |
ENST00000382171.2
|
KRTAP5-1
|
keratin associated protein 5-1 |
| chr15_+_81225699 | 0.03 |
ENST00000560027.1
|
KIAA1199
|
KIAA1199 |
| chr16_+_2802623 | 0.03 |
ENST00000576924.1
ENST00000575009.1 ENST00000576415.1 ENST00000571378.1 |
SRRM2
|
serine/arginine repetitive matrix 2 |
| chr10_-_96829246 | 0.03 |
ENST00000371270.3
ENST00000535898.1 ENST00000539050.1 |
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chr1_+_228337553 | 0.03 |
ENST00000366714.2
|
GJC2
|
gap junction protein, gamma 2, 47kDa |
| chr5_+_78532003 | 0.03 |
ENST00000396137.4
|
JMY
|
junction mediating and regulatory protein, p53 cofactor |
| chr1_-_20446020 | 0.03 |
ENST00000375105.3
|
PLA2G2D
|
phospholipase A2, group IID |
| chr8_+_11291429 | 0.03 |
ENST00000533578.1
|
C8orf12
|
chromosome 8 open reading frame 12 |
| chr12_-_90102594 | 0.03 |
ENST00000428670.3
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr10_+_76871454 | 0.03 |
ENST00000372687.4
|
SAMD8
|
sterile alpha motif domain containing 8 |
| chr13_-_79979952 | 0.03 |
ENST00000438724.1
|
RBM26
|
RNA binding motif protein 26 |
| chr1_-_114447412 | 0.03 |
ENST00000369567.1
ENST00000369566.3 |
AP4B1
|
adaptor-related protein complex 4, beta 1 subunit |
| chr16_-_65155833 | 0.02 |
ENST00000566827.1
ENST00000394156.3 ENST00000562998.1 |
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr11_+_56949221 | 0.02 |
ENST00000497933.1
|
LRRC55
|
leucine rich repeat containing 55 |
| chr8_+_32405728 | 0.02 |
ENST00000523079.1
ENST00000338921.4 ENST00000356819.4 ENST00000287845.5 ENST00000341377.5 |
NRG1
|
neuregulin 1 |
| chr11_-_45687128 | 0.02 |
ENST00000308064.2
|
CHST1
|
carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 |
| chr8_+_11351494 | 0.02 |
ENST00000259089.4
|
BLK
|
B lymphoid tyrosine kinase |
| chr16_+_2802316 | 0.02 |
ENST00000301740.8
|
SRRM2
|
serine/arginine repetitive matrix 2 |
| chr15_+_68924327 | 0.02 |
ENST00000543950.1
|
CORO2B
|
coronin, actin binding protein, 2B |
| chr14_+_70346125 | 0.02 |
ENST00000361956.3
ENST00000381280.4 |
SMOC1
|
SPARC related modular calcium binding 1 |
| chr1_+_178694300 | 0.02 |
ENST00000367635.3
|
RALGPS2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr12_-_120884175 | 0.02 |
ENST00000546954.1
|
TRIAP1
|
TP53 regulated inhibitor of apoptosis 1 |
| chr16_-_75569068 | 0.02 |
ENST00000336257.3
ENST00000565039.1 |
CHST5
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 5 |
| chr20_+_18125727 | 0.02 |
ENST00000489634.2
|
CSRP2BP
|
CSRP2 binding protein |
| chrX_-_21676442 | 0.02 |
ENST00000379499.2
|
KLHL34
|
kelch-like family member 34 |
| chr4_+_81951957 | 0.02 |
ENST00000282701.2
|
BMP3
|
bone morphogenetic protein 3 |
| chr3_-_122712079 | 0.02 |
ENST00000393583.2
|
SEMA5B
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5B |
| chr12_-_50677255 | 0.02 |
ENST00000551691.1
ENST00000394943.3 ENST00000341247.4 |
LIMA1
|
LIM domain and actin binding 1 |
| chr2_-_1748214 | 0.02 |
ENST00000433670.1
ENST00000425171.1 ENST00000252804.4 |
PXDN
|
peroxidasin homolog (Drosophila) |
| chr11_-_111794446 | 0.02 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr4_-_5990166 | 0.02 |
ENST00000324058.5
|
C4orf50
|
chromosome 4 open reading frame 50 |
| chr8_+_32405785 | 0.01 |
ENST00000287842.3
|
NRG1
|
neuregulin 1 |
| chr9_-_116061145 | 0.01 |
ENST00000416588.2
|
RNF183
|
ring finger protein 183 |
| chrX_-_13956737 | 0.01 |
ENST00000454189.2
|
GPM6B
|
glycoprotein M6B |
| chr1_-_33647267 | 0.01 |
ENST00000291416.5
|
TRIM62
|
tripartite motif containing 62 |
| chr9_+_33795533 | 0.01 |
ENST00000379405.3
|
PRSS3
|
protease, serine, 3 |
| chr22_-_50051151 | 0.01 |
ENST00000400023.1
ENST00000444628.1 |
C22orf34
|
chromosome 22 open reading frame 34 |
| chr13_-_79979919 | 0.01 |
ENST00000267229.7
|
RBM26
|
RNA binding motif protein 26 |
| chr20_+_44441215 | 0.01 |
ENST00000356455.4
ENST00000405520.1 |
UBE2C
|
ubiquitin-conjugating enzyme E2C |
| chr6_-_46620522 | 0.01 |
ENST00000275016.2
|
CYP39A1
|
cytochrome P450, family 39, subfamily A, polypeptide 1 |
| chr19_+_46801639 | 0.01 |
ENST00000244303.6
ENST00000339613.2 ENST00000533145.1 ENST00000472815.1 |
HIF3A
|
hypoxia inducible factor 3, alpha subunit |
| chr3_+_51663407 | 0.01 |
ENST00000432863.1
ENST00000296477.3 |
RAD54L2
|
RAD54-like 2 (S. cerevisiae) |
| chrX_+_15808569 | 0.01 |
ENST00000380308.3
ENST00000307771.7 |
ZRSR2
|
zinc finger (CCCH type), RNA-binding motif and serine/arginine rich 2 |
| chr1_-_9129598 | 0.01 |
ENST00000535586.1
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr6_-_56258892 | 0.01 |
ENST00000370819.1
|
COL21A1
|
collagen, type XXI, alpha 1 |
| chr2_-_237416071 | 0.01 |
ENST00000309507.5
ENST00000431676.2 |
IQCA1
|
IQ motif containing with AAA domain 1 |
| chr9_-_138853156 | 0.01 |
ENST00000371756.3
|
UBAC1
|
UBA domain containing 1 |
| chr7_-_99097863 | 0.01 |
ENST00000426306.2
ENST00000337673.6 |
ZNF394
|
zinc finger protein 394 |
| chr17_-_33416231 | 0.01 |
ENST00000584655.1
ENST00000447669.2 ENST00000315249.7 |
RFFL
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr2_+_183943464 | 0.01 |
ENST00000354221.4
|
DUSP19
|
dual specificity phosphatase 19 |
| chr12_+_113344755 | 0.00 |
ENST00000550883.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr19_+_29493456 | 0.00 |
ENST00000591143.1
ENST00000592653.1 |
CTD-2081K17.2
|
CTD-2081K17.2 |
| chr4_-_89744365 | 0.00 |
ENST00000513837.1
ENST00000503556.1 |
FAM13A
|
family with sequence similarity 13, member A |
| chr1_+_171454639 | 0.00 |
ENST00000392078.3
ENST00000426496.2 |
PRRC2C
|
proline-rich coiled-coil 2C |
| chr8_+_11351876 | 0.00 |
ENST00000529894.1
|
BLK
|
B lymphoid tyrosine kinase |
| chr7_+_150688166 | 0.00 |
ENST00000461406.1
|
NOS3
|
nitric oxide synthase 3 (endothelial cell) |
| chr2_-_237416181 | 0.00 |
ENST00000409907.3
|
IQCA1
|
IQ motif containing with AAA domain 1 |
| chr4_-_129491686 | 0.00 |
ENST00000514265.1
|
RP11-184M15.1
|
RP11-184M15.1 |
| chr10_+_123970670 | 0.00 |
ENST00000496913.2
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr19_-_40331345 | 0.00 |
ENST00000597224.1
|
FBL
|
fibrillarin |
| chr3_+_167453026 | 0.00 |
ENST00000472941.1
|
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr8_-_145641864 | 0.00 |
ENST00000276833.5
|
SLC39A4
|
solute carrier family 39 (zinc transporter), member 4 |
| chr1_+_26856236 | 0.00 |
ENST00000374168.2
ENST00000374166.4 |
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr3_-_88108212 | 0.00 |
ENST00000482016.1
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr4_-_175750364 | 0.00 |
ENST00000340217.5
ENST00000274093.3 |
GLRA3
|
glycine receptor, alpha 3 |
Network of associatons between targets according to the STRING database.
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | GO:1903179 | regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) |
| 0.1 | 0.2 | GO:0016999 | antibiotic metabolic process(GO:0016999) cellular amide catabolic process(GO:0043605) |
| 0.0 | 0.3 | GO:0097056 | selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.0 | 0.3 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.2 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.0 | 0.2 | GO:1903750 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.0 | 0.2 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.1 | GO:0039507 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.0 | 0.1 | GO:0032484 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.1 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.1 | GO:0031627 | telomeric loop formation(GO:0031627) telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.1 | GO:1902724 | mineralocorticoid receptor signaling pathway(GO:0031959) positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.0 | 0.0 | GO:0032759 | TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) |
| 0.0 | 0.1 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.1 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.0 | 0.0 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.1 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.0 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 0.0 | 0.1 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.3 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 0.1 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.0 | 0.3 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.0 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.1 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.1 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.0 | 0.1 | GO:0035240 | dopamine binding(GO:0035240) |
| 0.0 | 0.2 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.2 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.0 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |