Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
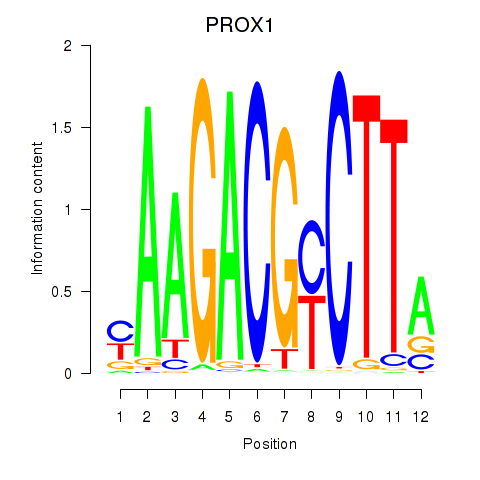
Results for PROX1
Z-value: 0.91
Transcription factors associated with PROX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PROX1
|
ENSG00000117707.11 | prospero homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PROX1 | hg19_v2_chr1_+_214161272_214161322 | -0.50 | 5.0e-01 | Click! |
Activity profile of PROX1 motif
Sorted Z-values of PROX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_142264664 | 0.84 |
ENST00000518520.1
|
RP11-10J21.3
|
Uncharacterized protein |
| chr6_-_8102279 | 0.71 |
ENST00000488226.2
|
EEF1E1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr21_-_34863693 | 0.57 |
ENST00000314399.3
|
DNAJC28
|
DnaJ (Hsp40) homolog, subfamily C, member 28 |
| chr10_+_32735030 | 0.52 |
ENST00000277657.6
ENST00000362006.5 |
CCDC7
|
coiled-coil domain containing 7 |
| chr6_-_107235331 | 0.46 |
ENST00000433965.1
ENST00000430094.1 |
RP1-60O19.1
|
RP1-60O19.1 |
| chr1_+_95975672 | 0.46 |
ENST00000440116.2
ENST00000456933.1 |
RP11-286B14.1
|
RP11-286B14.1 |
| chr12_+_42624050 | 0.39 |
ENST00000601185.1
|
AC020629.1
|
Uncharacterized protein |
| chr10_-_43633747 | 0.36 |
ENST00000609407.1
|
RP11-351D16.3
|
RP11-351D16.3 |
| chr21_-_26979786 | 0.34 |
ENST00000419219.1
ENST00000352957.4 ENST00000307301.7 |
MRPL39
|
mitochondrial ribosomal protein L39 |
| chr13_-_36788718 | 0.34 |
ENST00000317764.6
ENST00000379881.3 |
SOHLH2
|
spermatogenesis and oogenesis specific basic helix-loop-helix 2 |
| chr17_+_73629500 | 0.33 |
ENST00000375215.3
|
SMIM5
|
small integral membrane protein 5 |
| chr4_+_128886584 | 0.33 |
ENST00000513371.1
|
C4orf29
|
chromosome 4 open reading frame 29 |
| chr6_-_107235378 | 0.31 |
ENST00000606430.1
|
RP1-60O19.1
|
RP1-60O19.1 |
| chr10_+_16478942 | 0.30 |
ENST00000535784.2
ENST00000423462.2 ENST00000378000.1 |
PTER
|
phosphotriesterase related |
| chr13_+_37581115 | 0.30 |
ENST00000481013.1
|
EXOSC8
|
exosome component 8 |
| chr10_-_94301107 | 0.30 |
ENST00000436178.1
|
IDE
|
insulin-degrading enzyme |
| chr17_+_42081914 | 0.30 |
ENST00000293404.3
ENST00000589767.1 |
NAGS
|
N-acetylglutamate synthase |
| chr6_+_144665237 | 0.28 |
ENST00000421035.2
|
UTRN
|
utrophin |
| chr10_+_32735177 | 0.28 |
ENST00000545067.1
|
CCDC7
|
coiled-coil domain containing 7 |
| chr2_+_232826394 | 0.27 |
ENST00000409401.3
ENST00000441279.1 |
DIS3L2
|
DIS3 mitotic control homolog (S. cerevisiae)-like 2 |
| chr16_+_31724552 | 0.26 |
ENST00000539915.1
ENST00000316491.9 ENST00000399681.3 ENST00000398696.3 ENST00000534369.1 |
ZNF720
|
zinc finger protein 720 |
| chr4_-_103747011 | 0.26 |
ENST00000350435.7
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr1_-_154178803 | 0.26 |
ENST00000368525.3
|
C1orf189
|
chromosome 1 open reading frame 189 |
| chr14_-_35183755 | 0.26 |
ENST00000555765.1
|
CFL2
|
cofilin 2 (muscle) |
| chr11_+_93394805 | 0.25 |
ENST00000325212.6
ENST00000411936.1 ENST00000344196.4 |
KIAA1731
|
KIAA1731 |
| chr3_-_197024965 | 0.25 |
ENST00000392382.2
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr7_+_64254766 | 0.25 |
ENST00000307355.7
ENST00000359735.3 |
ZNF138
|
zinc finger protein 138 |
| chr2_-_163175133 | 0.24 |
ENST00000421365.2
ENST00000263642.2 |
IFIH1
|
interferon induced with helicase C domain 1 |
| chr6_-_159420780 | 0.24 |
ENST00000449822.1
|
RSPH3
|
radial spoke 3 homolog (Chlamydomonas) |
| chr10_-_14920599 | 0.24 |
ENST00000609399.1
|
RP11-398C13.6
|
RP11-398C13.6 |
| chr1_+_154909803 | 0.24 |
ENST00000604546.1
|
RP11-307C12.13
|
RP11-307C12.13 |
| chr11_+_111749650 | 0.23 |
ENST00000528125.1
|
C11orf1
|
chromosome 11 open reading frame 1 |
| chr7_+_64254793 | 0.23 |
ENST00000494380.1
ENST00000440155.2 ENST00000440598.1 ENST00000437743.1 |
ZNF138
|
zinc finger protein 138 |
| chr21_-_16125773 | 0.23 |
ENST00000454128.2
|
AF127936.3
|
AF127936.3 |
| chr17_+_41994576 | 0.23 |
ENST00000588043.2
|
FAM215A
|
family with sequence similarity 215, member A (non-protein coding) |
| chr4_-_169931393 | 0.23 |
ENST00000504480.1
ENST00000306193.3 |
CBR4
|
carbonyl reductase 4 |
| chr4_-_170947446 | 0.22 |
ENST00000507601.1
ENST00000512698.1 |
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr10_+_30723045 | 0.22 |
ENST00000542547.1
ENST00000415139.1 |
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr7_+_156433363 | 0.21 |
ENST00000439609.1
|
RNF32
|
ring finger protein 32 |
| chr10_+_112327425 | 0.20 |
ENST00000361804.4
|
SMC3
|
structural maintenance of chromosomes 3 |
| chr4_-_170948361 | 0.20 |
ENST00000393702.3
|
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr8_+_110552046 | 0.20 |
ENST00000529931.1
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9 |
| chr1_+_171454639 | 0.20 |
ENST00000392078.3
ENST00000426496.2 |
PRRC2C
|
proline-rich coiled-coil 2C |
| chr7_+_92158083 | 0.20 |
ENST00000265732.5
ENST00000481551.1 ENST00000496410.1 |
RBM48
|
RNA binding motif protein 48 |
| chr4_-_170947522 | 0.19 |
ENST00000361618.3
|
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr4_+_128886532 | 0.19 |
ENST00000444616.1
ENST00000388795.5 |
C4orf29
|
chromosome 4 open reading frame 29 |
| chr9_-_80437915 | 0.19 |
ENST00000397476.3
|
GNAQ
|
guanine nucleotide binding protein (G protein), q polypeptide |
| chr2_-_107084826 | 0.19 |
ENST00000304514.7
ENST00000409886.3 |
RGPD3
|
RANBP2-like and GRIP domain containing 3 |
| chr19_-_40732594 | 0.19 |
ENST00000430325.2
ENST00000433940.1 |
CNTD2
|
cyclin N-terminal domain containing 2 |
| chr1_+_35544968 | 0.19 |
ENST00000359858.4
ENST00000373330.1 |
ZMYM1
|
zinc finger, MYM-type 1 |
| chr10_+_14920843 | 0.18 |
ENST00000433779.1
ENST00000378325.3 ENST00000354919.6 ENST00000313519.5 ENST00000420416.1 |
SUV39H2
|
suppressor of variegation 3-9 homolog 2 (Drosophila) |
| chr1_+_179263308 | 0.18 |
ENST00000426956.1
|
SOAT1
|
sterol O-acyltransferase 1 |
| chr3_-_96337000 | 0.17 |
ENST00000600213.2
|
MTRNR2L12
|
MT-RNR2-like 12 (pseudogene) |
| chr4_-_159592996 | 0.17 |
ENST00000508457.1
|
C4orf46
|
chromosome 4 open reading frame 46 |
| chr5_-_156569850 | 0.17 |
ENST00000524219.1
|
HAVCR2
|
hepatitis A virus cellular receptor 2 |
| chr15_+_85523671 | 0.17 |
ENST00000310298.4
ENST00000557957.1 |
PDE8A
|
phosphodiesterase 8A |
| chr3_+_12525931 | 0.17 |
ENST00000446004.1
ENST00000314571.7 ENST00000454502.2 ENST00000383797.5 ENST00000402228.3 ENST00000284995.6 ENST00000444864.1 |
TSEN2
|
TSEN2 tRNA splicing endonuclease subunit |
| chr1_+_70876891 | 0.17 |
ENST00000411986.2
|
CTH
|
cystathionase (cystathionine gamma-lyase) |
| chr10_-_75385711 | 0.17 |
ENST00000433394.1
|
USP54
|
ubiquitin specific peptidase 54 |
| chr3_+_136676707 | 0.17 |
ENST00000329582.4
|
IL20RB
|
interleukin 20 receptor beta |
| chr5_+_131892603 | 0.17 |
ENST00000378823.3
ENST00000265335.6 |
RAD50
|
RAD50 homolog (S. cerevisiae) |
| chr8_-_135652051 | 0.16 |
ENST00000522257.1
|
ZFAT
|
zinc finger and AT hook domain containing |
| chr15_-_101835110 | 0.16 |
ENST00000560496.1
|
SNRPA1
|
small nuclear ribonucleoprotein polypeptide A' |
| chr4_-_103746924 | 0.16 |
ENST00000505207.1
ENST00000502404.1 ENST00000507845.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr9_+_72873837 | 0.16 |
ENST00000361138.5
|
SMC5
|
structural maintenance of chromosomes 5 |
| chr21_-_16126181 | 0.16 |
ENST00000455253.2
|
AF127936.3
|
AF127936.3 |
| chr2_+_131862900 | 0.16 |
ENST00000438882.2
ENST00000538982.1 ENST00000404460.1 |
PLEKHB2
|
pleckstrin homology domain containing, family B (evectins) member 2 |
| chr11_+_61722629 | 0.16 |
ENST00000526988.1
|
BEST1
|
bestrophin 1 |
| chr2_+_131862872 | 0.16 |
ENST00000439822.2
|
PLEKHB2
|
pleckstrin homology domain containing, family B (evectins) member 2 |
| chr13_+_49822041 | 0.15 |
ENST00000538056.1
ENST00000251108.6 ENST00000444959.1 ENST00000429346.1 |
CDADC1
|
cytidine and dCMP deaminase domain containing 1 |
| chr16_+_28857957 | 0.15 |
ENST00000567536.1
|
SH2B1
|
SH2B adaptor protein 1 |
| chr6_-_72129806 | 0.15 |
ENST00000413945.1
ENST00000602878.1 ENST00000436803.1 ENST00000421704.1 ENST00000441570.1 |
LINC00472
|
long intergenic non-protein coding RNA 472 |
| chr1_+_104068562 | 0.15 |
ENST00000423855.2
|
RNPC3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr3_+_138067314 | 0.15 |
ENST00000423968.2
|
MRAS
|
muscle RAS oncogene homolog |
| chr10_+_71075516 | 0.15 |
ENST00000436817.1
|
HK1
|
hexokinase 1 |
| chr3_-_197025447 | 0.15 |
ENST00000346964.2
ENST00000357674.4 ENST00000314062.3 ENST00000448528.2 ENST00000419553.1 |
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr15_-_78913521 | 0.15 |
ENST00000326828.5
|
CHRNA3
|
cholinergic receptor, nicotinic, alpha 3 (neuronal) |
| chr11_-_73882057 | 0.15 |
ENST00000334126.7
ENST00000313663.7 |
C2CD3
|
C2 calcium-dependent domain containing 3 |
| chr17_+_16593539 | 0.15 |
ENST00000340621.5
ENST00000399273.1 ENST00000443444.2 ENST00000360524.8 ENST00000456009.1 |
CCDC144A
|
coiled-coil domain containing 144A |
| chr7_-_143105941 | 0.15 |
ENST00000275815.3
|
EPHA1
|
EPH receptor A1 |
| chr11_+_111749942 | 0.15 |
ENST00000260276.3
|
C11orf1
|
chromosome 11 open reading frame 1 |
| chr10_+_30723105 | 0.14 |
ENST00000375322.2
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr1_+_179335101 | 0.14 |
ENST00000508285.1
ENST00000511889.1 |
AXDND1
|
axonemal dynein light chain domain containing 1 |
| chr18_-_2571210 | 0.14 |
ENST00000577166.1
|
METTL4
|
methyltransferase like 4 |
| chr5_+_145583107 | 0.14 |
ENST00000506502.1
|
RBM27
|
RNA binding motif protein 27 |
| chr2_-_65659762 | 0.14 |
ENST00000440972.1
|
SPRED2
|
sprouty-related, EVH1 domain containing 2 |
| chr7_-_91509986 | 0.14 |
ENST00000456229.1
ENST00000442961.1 ENST00000406735.2 ENST00000419292.1 ENST00000351870.3 |
MTERF
|
mitochondrial transcription termination factor |
| chr1_-_28241024 | 0.14 |
ENST00000313433.7
ENST00000444045.1 |
RPA2
|
replication protein A2, 32kDa |
| chr5_+_64859066 | 0.14 |
ENST00000261308.5
ENST00000535264.1 ENST00000538977.1 |
PPWD1
|
peptidylprolyl isomerase domain and WD repeat containing 1 |
| chr5_+_145826867 | 0.14 |
ENST00000296702.5
ENST00000394421.2 |
TCERG1
|
transcription elongation regulator 1 |
| chr1_+_93297582 | 0.13 |
ENST00000370321.3
|
RPL5
|
ribosomal protein L5 |
| chr4_-_103746683 | 0.13 |
ENST00000504211.1
ENST00000508476.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chrX_+_117861535 | 0.13 |
ENST00000371666.3
ENST00000371642.1 |
IL13RA1
|
interleukin 13 receptor, alpha 1 |
| chr12_-_66524482 | 0.13 |
ENST00000446587.2
ENST00000266604.2 |
LLPH
|
LLP homolog, long-term synaptic facilitation (Aplysia) |
| chr19_+_35773242 | 0.13 |
ENST00000222304.3
|
HAMP
|
hepcidin antimicrobial peptide |
| chr19_-_9546177 | 0.13 |
ENST00000592292.1
ENST00000588221.1 |
ZNF266
|
zinc finger protein 266 |
| chr2_-_225434538 | 0.13 |
ENST00000409096.1
|
CUL3
|
cullin 3 |
| chr17_+_66243715 | 0.12 |
ENST00000359904.3
|
AMZ2
|
archaelysin family metallopeptidase 2 |
| chr2_+_108443388 | 0.12 |
ENST00000354986.4
ENST00000408999.3 |
RGPD4
|
RANBP2-like and GRIP domain containing 4 |
| chr16_+_23194033 | 0.12 |
ENST00000300061.2
|
SCNN1G
|
sodium channel, non-voltage-gated 1, gamma subunit |
| chr1_+_70876926 | 0.12 |
ENST00000370938.3
ENST00000346806.2 |
CTH
|
cystathionase (cystathionine gamma-lyase) |
| chr7_-_143582450 | 0.12 |
ENST00000485416.1
|
FAM115A
|
family with sequence similarity 115, member A |
| chr7_-_25164868 | 0.12 |
ENST00000409409.1
ENST00000409764.1 ENST00000413447.1 |
CYCS
|
cytochrome c, somatic |
| chr9_+_131218698 | 0.12 |
ENST00000434106.3
ENST00000546203.1 ENST00000446274.1 ENST00000421776.2 ENST00000432065.2 |
ODF2
|
outer dense fiber of sperm tails 2 |
| chrX_+_150565038 | 0.11 |
ENST00000370361.1
|
VMA21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chr19_-_9546227 | 0.11 |
ENST00000361451.2
ENST00000361151.1 |
ZNF266
|
zinc finger protein 266 |
| chr11_-_46408107 | 0.11 |
ENST00000433765.2
|
CHRM4
|
cholinergic receptor, muscarinic 4 |
| chr12_+_121078355 | 0.11 |
ENST00000316803.3
|
CABP1
|
calcium binding protein 1 |
| chr5_+_145583156 | 0.11 |
ENST00000265271.5
|
RBM27
|
RNA binding motif protein 27 |
| chr19_+_6361754 | 0.11 |
ENST00000597326.1
|
CLPP
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr5_+_154320623 | 0.11 |
ENST00000523037.1
ENST00000265229.8 ENST00000439747.3 ENST00000522038.1 |
MRPL22
|
mitochondrial ribosomal protein L22 |
| chr3_-_196014520 | 0.11 |
ENST00000441879.1
ENST00000292823.2 ENST00000411591.1 ENST00000431016.1 ENST00000443555.1 |
PCYT1A
|
phosphate cytidylyltransferase 1, choline, alpha |
| chr17_+_55183261 | 0.11 |
ENST00000576295.1
|
AKAP1
|
A kinase (PRKA) anchor protein 1 |
| chr2_-_54087066 | 0.11 |
ENST00000394705.2
ENST00000352846.3 ENST00000406625.2 |
GPR75
GPR75-ASB3
ASB3
|
G protein-coupled receptor 75 GPR75-ASB3 readthrough Ankyrin repeat and SOCS box protein 3 |
| chr18_+_2571510 | 0.11 |
ENST00000261597.4
ENST00000575515.1 |
NDC80
|
NDC80 kinetochore complex component |
| chr11_+_73360024 | 0.11 |
ENST00000540431.1
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr10_-_121296045 | 0.10 |
ENST00000392865.1
|
RGS10
|
regulator of G-protein signaling 10 |
| chr10_+_30722866 | 0.10 |
ENST00000263056.1
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr2_+_97001491 | 0.10 |
ENST00000240423.4
ENST00000427946.1 ENST00000435975.1 ENST00000456906.1 ENST00000455200.1 |
NCAPH
|
non-SMC condensin I complex, subunit H |
| chr5_+_179125368 | 0.10 |
ENST00000502296.1
ENST00000504734.1 ENST00000415618.2 |
CANX
|
calnexin |
| chr12_+_113229737 | 0.10 |
ENST00000551052.1
ENST00000415485.3 |
RPH3A
|
rabphilin 3A homolog (mouse) |
| chr7_-_91509972 | 0.10 |
ENST00000425936.1
|
MTERF
|
mitochondrial transcription termination factor |
| chr4_-_169931231 | 0.10 |
ENST00000504561.1
|
CBR4
|
carbonyl reductase 4 |
| chr1_+_159770292 | 0.10 |
ENST00000536257.1
ENST00000321935.6 |
FCRL6
|
Fc receptor-like 6 |
| chr15_-_91537723 | 0.10 |
ENST00000394249.3
ENST00000559811.1 ENST00000442656.2 ENST00000557905.1 ENST00000361919.3 |
PRC1
|
protein regulator of cytokinesis 1 |
| chr4_-_140477353 | 0.10 |
ENST00000406354.1
ENST00000506866.2 |
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr8_-_146176199 | 0.10 |
ENST00000532351.1
ENST00000276816.4 ENST00000394909.2 |
ZNF16
|
zinc finger protein 16 |
| chr17_+_38337491 | 0.10 |
ENST00000538981.1
|
RAPGEFL1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr20_+_57209828 | 0.10 |
ENST00000598340.1
|
MGC4294
|
HCG1785561; MGC4294 protein; Uncharacterized protein |
| chr5_-_132113063 | 0.10 |
ENST00000378719.2
|
SEPT8
|
septin 8 |
| chr12_-_11091862 | 0.09 |
ENST00000537503.1
|
TAS2R14
|
taste receptor, type 2, member 14 |
| chr19_-_15529790 | 0.09 |
ENST00000596195.1
ENST00000595067.1 ENST00000595465.2 ENST00000397410.5 ENST00000600247.1 |
AKAP8L
|
A kinase (PRKA) anchor protein 8-like |
| chr1_+_104068312 | 0.09 |
ENST00000524631.1
ENST00000531883.1 ENST00000533099.1 ENST00000527062.1 |
RNPC3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr16_+_31885079 | 0.09 |
ENST00000300870.10
ENST00000394846.3 |
ZNF267
|
zinc finger protein 267 |
| chr10_+_60028818 | 0.09 |
ENST00000333926.5
|
CISD1
|
CDGSH iron sulfur domain 1 |
| chr10_-_92681033 | 0.09 |
ENST00000371697.3
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle) |
| chr6_+_126102292 | 0.09 |
ENST00000368357.3
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr16_-_49890016 | 0.09 |
ENST00000563137.2
|
ZNF423
|
zinc finger protein 423 |
| chr1_+_82266053 | 0.09 |
ENST00000370715.1
ENST00000370713.1 ENST00000319517.6 ENST00000370717.2 ENST00000394879.1 ENST00000271029.4 ENST00000335786.5 |
LPHN2
|
latrophilin 2 |
| chr11_+_64009072 | 0.09 |
ENST00000535135.1
ENST00000394540.3 |
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr9_+_131266963 | 0.09 |
ENST00000309971.4
ENST00000372770.4 |
GLE1
|
GLE1 RNA export mediator |
| chr3_-_122283424 | 0.09 |
ENST00000477522.2
ENST00000360356.2 |
PARP9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr1_+_161719552 | 0.09 |
ENST00000367943.4
|
DUSP12
|
dual specificity phosphatase 12 |
| chr5_+_6766004 | 0.08 |
ENST00000506093.1
|
RP11-332J15.3
|
RP11-332J15.3 |
| chr8_+_110552831 | 0.08 |
ENST00000530629.1
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9 |
| chr15_+_38746307 | 0.08 |
ENST00000397609.2
ENST00000491535.1 |
FAM98B
|
family with sequence similarity 98, member B |
| chr11_+_117947724 | 0.08 |
ENST00000534111.1
|
TMPRSS4
|
transmembrane protease, serine 4 |
| chr7_+_66093908 | 0.08 |
ENST00000443322.1
ENST00000449064.1 |
KCTD7
|
potassium channel tetramerization domain containing 7 |
| chr10_+_134258649 | 0.08 |
ENST00000392630.3
ENST00000321248.2 |
C10orf91
|
chromosome 10 open reading frame 91 |
| chr10_-_52008313 | 0.08 |
ENST00000329428.6
ENST00000395526.4 ENST00000447815.1 |
ASAH2
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2 |
| chr1_+_10459111 | 0.08 |
ENST00000541529.1
ENST00000270776.8 ENST00000483936.1 ENST00000538557.1 |
PGD
|
phosphogluconate dehydrogenase |
| chr16_-_22012419 | 0.08 |
ENST00000537222.2
ENST00000424898.2 ENST00000286143.6 |
PDZD9
|
PDZ domain containing 9 |
| chr4_+_128886424 | 0.08 |
ENST00000398965.1
|
C4orf29
|
chromosome 4 open reading frame 29 |
| chr6_-_74104856 | 0.08 |
ENST00000441145.1
|
OOEP
|
oocyte expressed protein |
| chr6_-_90529418 | 0.08 |
ENST00000439638.1
ENST00000369393.3 ENST00000428876.1 |
MDN1
|
MDN1, midasin homolog (yeast) |
| chr4_+_26862313 | 0.08 |
ENST00000467087.1
ENST00000382009.3 ENST00000237364.5 |
STIM2
|
stromal interaction molecule 2 |
| chr9_+_132096166 | 0.08 |
ENST00000436710.1
|
RP11-65J3.1
|
RP11-65J3.1 |
| chr10_+_71075552 | 0.07 |
ENST00000298649.3
|
HK1
|
hexokinase 1 |
| chr19_+_58281014 | 0.07 |
ENST00000391702.3
ENST00000598885.1 ENST00000598183.1 ENST00000396154.2 ENST00000599802.1 ENST00000396150.4 |
ZNF586
|
zinc finger protein 586 |
| chr5_+_156569944 | 0.07 |
ENST00000521769.1
|
ITK
|
IL2-inducible T-cell kinase |
| chr11_-_82997371 | 0.07 |
ENST00000525503.1
|
CCDC90B
|
coiled-coil domain containing 90B |
| chr11_+_64008525 | 0.07 |
ENST00000449942.2
|
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr1_+_203830703 | 0.07 |
ENST00000414487.2
|
SNRPE
|
small nuclear ribonucleoprotein polypeptide E |
| chr14_-_105708942 | 0.07 |
ENST00000549655.1
|
BRF1
|
BRF1, RNA polymerase III transcription initiation factor 90 kDa subunit |
| chr19_+_15752088 | 0.07 |
ENST00000585846.1
|
CYP4F3
|
cytochrome P450, family 4, subfamily F, polypeptide 3 |
| chr2_-_42588338 | 0.07 |
ENST00000234301.2
|
COX7A2L
|
cytochrome c oxidase subunit VIIa polypeptide 2 like |
| chr1_-_109203685 | 0.07 |
ENST00000402983.1
ENST00000420055.1 |
HENMT1
|
HEN1 methyltransferase homolog 1 (Arabidopsis) |
| chr1_+_171454659 | 0.07 |
ENST00000367742.3
ENST00000338920.4 |
PRRC2C
|
proline-rich coiled-coil 2C |
| chr8_-_13134045 | 0.07 |
ENST00000512044.2
|
DLC1
|
deleted in liver cancer 1 |
| chrX_+_70586140 | 0.07 |
ENST00000276072.3
|
TAF1
|
TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250kDa |
| chr2_+_139259324 | 0.07 |
ENST00000280098.4
|
SPOPL
|
speckle-type POZ protein-like |
| chr16_-_21442874 | 0.07 |
ENST00000534903.1
|
NPIPB3
|
nuclear pore complex interacting protein family, member B3 |
| chr2_+_17935383 | 0.06 |
ENST00000524465.1
ENST00000381254.2 ENST00000532257.1 |
GEN1
|
GEN1 Holliday junction 5' flap endonuclease |
| chr4_+_26862431 | 0.06 |
ENST00000465503.1
|
STIM2
|
stromal interaction molecule 2 |
| chr1_-_202311088 | 0.06 |
ENST00000367274.4
|
UBE2T
|
ubiquitin-conjugating enzyme E2T (putative) |
| chr2_+_101179152 | 0.06 |
ENST00000264254.6
|
PDCL3
|
phosducin-like 3 |
| chr4_-_69215467 | 0.06 |
ENST00000579690.1
|
YTHDC1
|
YTH domain containing 1 |
| chr14_-_68141535 | 0.06 |
ENST00000554659.1
|
VTI1B
|
vesicle transport through interaction with t-SNAREs 1B |
| chr11_-_59383617 | 0.06 |
ENST00000263847.1
|
OSBP
|
oxysterol binding protein |
| chr20_-_749121 | 0.06 |
ENST00000217254.7
ENST00000381944.3 |
SLC52A3
|
solute carrier family 52 (riboflavin transporter), member 3 |
| chr10_+_45869652 | 0.06 |
ENST00000542434.1
ENST00000374391.2 |
ALOX5
|
arachidonate 5-lipoxygenase |
| chr7_-_25164969 | 0.06 |
ENST00000305786.2
|
CYCS
|
cytochrome c, somatic |
| chr10_-_96122682 | 0.06 |
ENST00000371361.3
|
NOC3L
|
nucleolar complex associated 3 homolog (S. cerevisiae) |
| chr11_-_82997013 | 0.05 |
ENST00000529073.1
ENST00000529611.1 |
CCDC90B
|
coiled-coil domain containing 90B |
| chr4_+_26862400 | 0.05 |
ENST00000467011.1
ENST00000412829.2 |
STIM2
|
stromal interaction molecule 2 |
| chr14_-_31676964 | 0.05 |
ENST00000553700.1
|
HECTD1
|
HECT domain containing E3 ubiquitin protein ligase 1 |
| chr15_+_59279851 | 0.05 |
ENST00000348370.4
ENST00000434298.1 ENST00000559160.1 |
RNF111
|
ring finger protein 111 |
| chr8_-_37824442 | 0.05 |
ENST00000345060.3
|
ADRB3
|
adrenoceptor beta 3 |
| chr14_+_73603126 | 0.05 |
ENST00000557356.1
ENST00000556864.1 ENST00000556533.1 ENST00000556951.1 ENST00000557293.1 ENST00000553719.1 ENST00000553599.1 ENST00000556011.1 ENST00000394157.3 ENST00000357710.4 ENST00000324501.5 ENST00000560005.2 ENST00000555254.1 ENST00000261970.3 ENST00000344094.3 ENST00000554131.1 ENST00000557037.1 |
PSEN1
|
presenilin 1 |
| chr9_+_93564191 | 0.05 |
ENST00000375747.1
|
SYK
|
spleen tyrosine kinase |
| chr17_+_66539369 | 0.05 |
ENST00000600820.1
|
AC079210.1
|
Uncharacterized protein; cDNA FLJ45097 fis, clone BRAWH3031054 |
| chr6_+_170151708 | 0.05 |
ENST00000592367.1
ENST00000590711.1 ENST00000366772.2 ENST00000592745.1 ENST00000392095.4 ENST00000366773.3 ENST00000586341.1 ENST00000418781.3 ENST00000588437.1 |
ERMARD
|
ER membrane-associated RNA degradation |
| chr3_+_179040767 | 0.05 |
ENST00000496856.1
ENST00000491818.1 |
ZNF639
|
zinc finger protein 639 |
| chr14_+_22788560 | 0.05 |
ENST00000390468.1
|
TRAV41
|
T cell receptor alpha variable 41 |
| chr1_-_118727781 | 0.05 |
ENST00000336338.5
|
SPAG17
|
sperm associated antigen 17 |
| chr6_+_170151887 | 0.05 |
ENST00000588451.1
|
ERMARD
|
ER membrane-associated RNA degradation |
| chr2_+_128403720 | 0.05 |
ENST00000272644.3
|
GPR17
|
G protein-coupled receptor 17 |
| chr11_-_64739358 | 0.05 |
ENST00000301896.5
ENST00000530444.1 |
C11orf85
|
chromosome 11 open reading frame 85 |
| chr12_+_113229543 | 0.05 |
ENST00000447659.2
|
RPH3A
|
rabphilin 3A homolog (mouse) |
| chr11_+_1874200 | 0.05 |
ENST00000311604.3
|
LSP1
|
lymphocyte-specific protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PROX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.1 | 0.2 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.1 | 0.2 | GO:0002835 | negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) negative regulation of natural killer cell mediated immune response to tumor cell(GO:0002856) negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002859) |
| 0.1 | 0.3 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.1 | 0.3 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.1 | 0.3 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.1 | 0.2 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.1 | 0.7 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.1 | GO:1990641 | cellular response to bile acid(GO:1903413) response to iron ion starvation(GO:1990641) |
| 0.0 | 0.2 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.2 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.0 | 0.4 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.3 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.0 | 0.2 | GO:0034093 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.3 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.2 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.1 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.3 | GO:0044598 | polyketide metabolic process(GO:0030638) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 | 0.2 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.1 | GO:0071140 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.0 | 0.2 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.1 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.2 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.0 | 0.1 | GO:0019521 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.2 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.2 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.0 | 0.3 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.3 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.1 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 | 0.1 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.0 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.1 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.1 | GO:0036369 | transcription factor catabolic process(GO:0036369) |
| 0.0 | 0.0 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.0 | 0.1 | GO:0032752 | response to molecule of fungal origin(GO:0002238) serotonin production involved in inflammatory response(GO:0002351) serotonin secretion involved in inflammatory response(GO:0002442) serotonin secretion by platelet(GO:0002554) positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) cellular response to molecule of fungal origin(GO:0071226) |
| 0.0 | 0.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.1 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.1 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.1 | GO:2000373 | regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.0 | 0.2 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.2 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.7 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.6 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.4 | GO:0097025 | lateral loop(GO:0043219) MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.2 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.4 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.1 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.1 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.0 | 0.0 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.3 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.3 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.2 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0003955 | NAD(P)H dehydrogenase (quinone) activity(GO:0003955) 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.1 | 0.3 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.1 | 0.2 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.1 | 0.2 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.1 | 0.3 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 0.2 | GO:0045155 | electron transporter, transferring electrons from CoQH2-cytochrome c reductase complex and cytochrome c oxidase complex activity(GO:0045155) |
| 0.1 | 0.2 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.2 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.4 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.2 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.2 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.2 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.2 | GO:1904047 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) S-adenosyl-L-methionine binding(GO:1904047) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.3 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0052871 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0001026 | TFIIIB-type transcription factor activity(GO:0001026) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.2 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.3 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.1 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.0 | 0.1 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.0 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.0 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.1 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.5 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.7 | PID BARD1 PATHWAY | BARD1 signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.7 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.6 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.2 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |