Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
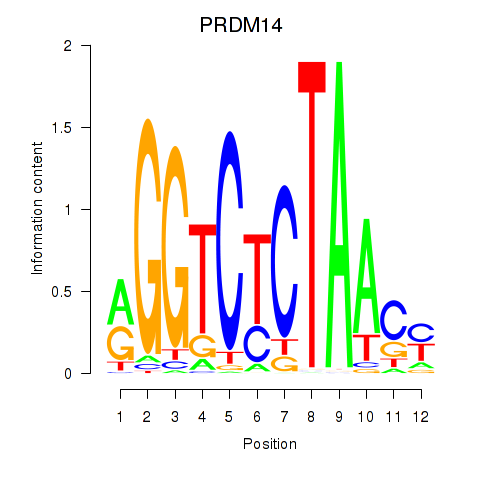
Results for PRDM14
Z-value: 0.25
Transcription factors associated with PRDM14
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PRDM14
|
ENSG00000147596.3 | PR/SET domain 14 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PRDM14 | hg19_v2_chr8_-_70983506_70983562 | 0.06 | 9.4e-01 | Click! |
Activity profile of PRDM14 motif
Sorted Z-values of PRDM14 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_134092561 | 0.12 |
ENST00000510560.1
ENST00000504234.1 ENST00000515172.1 |
AMOTL2
|
angiomotin like 2 |
| chr17_-_19651598 | 0.08 |
ENST00000570414.1
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr19_+_49838653 | 0.07 |
ENST00000598095.1
ENST00000426897.2 ENST00000323906.4 ENST00000535669.2 ENST00000597602.1 ENST00000595660.1 |
CD37
|
CD37 molecule |
| chr15_-_45694380 | 0.05 |
ENST00000561148.1
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr5_-_108063949 | 0.05 |
ENST00000606054.1
|
LINC01023
|
long intergenic non-protein coding RNA 1023 |
| chr4_-_10459009 | 0.05 |
ENST00000507515.1
|
ZNF518B
|
zinc finger protein 518B |
| chr8_+_101349823 | 0.05 |
ENST00000519566.1
|
KB-1991G8.1
|
KB-1991G8.1 |
| chr17_-_17726907 | 0.05 |
ENST00000423161.3
|
SREBF1
|
sterol regulatory element binding transcription factor 1 |
| chr11_+_64879317 | 0.04 |
ENST00000526809.1
ENST00000279263.7 ENST00000524986.1 ENST00000534371.1 ENST00000540748.1 ENST00000525385.1 ENST00000345348.5 ENST00000531321.1 ENST00000529414.1 ENST00000526085.1 ENST00000530750.1 |
TM7SF2
|
transmembrane 7 superfamily member 2 |
| chr17_-_19651654 | 0.04 |
ENST00000395555.3
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr11_-_46615498 | 0.04 |
ENST00000533727.1
ENST00000534300.1 ENST00000528950.1 ENST00000526606.1 |
AMBRA1
|
autophagy/beclin-1 regulator 1 |
| chr1_-_155006224 | 0.04 |
ENST00000368424.3
|
DCST2
|
DC-STAMP domain containing 2 |
| chr13_-_34250861 | 0.04 |
ENST00000445227.1
ENST00000454681.2 |
RP11-141M1.3
|
RP11-141M1.3 |
| chr1_-_155006254 | 0.04 |
ENST00000295536.5
|
DCST2
|
DC-STAMP domain containing 2 |
| chr17_-_2117600 | 0.04 |
ENST00000572369.1
|
SMG6
|
SMG6 nonsense mediated mRNA decay factor |
| chr20_-_3154162 | 0.03 |
ENST00000360342.3
|
LZTS3
|
Homo sapiens leucine zipper, putative tumor suppressor family member 3 (LZTS3), mRNA. |
| chr17_+_36873677 | 0.03 |
ENST00000471200.1
|
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6 |
| chr3_-_47517302 | 0.03 |
ENST00000441517.2
ENST00000545718.1 |
SCAP
|
SREBF chaperone |
| chr12_-_49351303 | 0.03 |
ENST00000256682.4
|
ARF3
|
ADP-ribosylation factor 3 |
| chr17_-_17485731 | 0.03 |
ENST00000395783.1
|
PEMT
|
phosphatidylethanolamine N-methyltransferase |
| chr12_+_56325812 | 0.03 |
ENST00000394147.1
ENST00000551156.1 ENST00000553783.1 ENST00000557080.1 ENST00000432422.3 ENST00000556001.1 |
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr15_+_45694567 | 0.03 |
ENST00000559860.1
|
SPATA5L1
|
spermatogenesis associated 5-like 1 |
| chr17_-_19651668 | 0.03 |
ENST00000494157.2
ENST00000225740.6 |
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr12_-_56326402 | 0.03 |
ENST00000547925.1
|
WIBG
|
within bgcn homolog (Drosophila) |
| chr1_+_45274154 | 0.03 |
ENST00000450269.1
ENST00000453418.1 ENST00000409335.2 |
BTBD19
|
BTB (POZ) domain containing 19 |
| chr1_+_36621529 | 0.03 |
ENST00000316156.4
|
MAP7D1
|
MAP7 domain containing 1 |
| chr2_-_208030295 | 0.03 |
ENST00000458272.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr6_+_43140095 | 0.03 |
ENST00000457278.2
|
SRF
|
serum response factor (c-fos serum response element-binding transcription factor) |
| chr4_+_4861385 | 0.03 |
ENST00000382723.4
|
MSX1
|
msh homeobox 1 |
| chr7_-_124405681 | 0.03 |
ENST00000303921.2
|
GPR37
|
G protein-coupled receptor 37 (endothelin receptor type B-like) |
| chr1_+_155006300 | 0.02 |
ENST00000295542.1
ENST00000392480.1 ENST00000423025.2 ENST00000368419.2 |
DCST1
|
DC-STAMP domain containing 1 |
| chr19_-_49314269 | 0.02 |
ENST00000545387.2
ENST00000316273.6 ENST00000402551.1 ENST00000598162.1 ENST00000599246.1 |
BCAT2
|
branched chain amino-acid transaminase 2, mitochondrial |
| chr3_-_155394099 | 0.02 |
ENST00000414191.1
|
PLCH1
|
phospholipase C, eta 1 |
| chr9_+_132597722 | 0.02 |
ENST00000372429.3
ENST00000315480.4 ENST00000358355.1 |
USP20
|
ubiquitin specific peptidase 20 |
| chr6_-_109777128 | 0.02 |
ENST00000358807.3
ENST00000358577.3 |
MICAL1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr1_+_27114418 | 0.02 |
ENST00000078527.4
|
PIGV
|
phosphatidylinositol glycan anchor biosynthesis, class V |
| chr5_+_32710736 | 0.02 |
ENST00000415685.2
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr1_+_27114589 | 0.02 |
ENST00000431541.1
ENST00000449950.2 ENST00000374145.1 |
PIGV
|
phosphatidylinositol glycan anchor biosynthesis, class V |
| chrX_+_51636629 | 0.02 |
ENST00000375722.1
ENST00000326587.7 ENST00000375695.2 |
MAGED1
|
melanoma antigen family D, 1 |
| chr11_+_62475130 | 0.02 |
ENST00000294117.5
|
GNG3
|
guanine nucleotide binding protein (G protein), gamma 3 |
| chr4_+_141677577 | 0.02 |
ENST00000609937.1
|
RP11-102N12.3
|
RP11-102N12.3 |
| chr3_-_155394152 | 0.01 |
ENST00000494598.1
|
PLCH1
|
phospholipase C, eta 1 |
| chr11_-_62474803 | 0.01 |
ENST00000533982.1
ENST00000360796.5 |
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr2_-_228244013 | 0.01 |
ENST00000304568.3
|
TM4SF20
|
transmembrane 4 L six family member 20 |
| chr17_+_41924536 | 0.01 |
ENST00000317310.4
|
CD300LG
|
CD300 molecule-like family member g |
| chr7_-_139763521 | 0.01 |
ENST00000263549.3
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr3_+_10289707 | 0.01 |
ENST00000287652.4
|
TATDN2
|
TatD DNase domain containing 2 |
| chr11_+_85566422 | 0.01 |
ENST00000342404.3
|
CCDC83
|
coiled-coil domain containing 83 |
| chr2_+_113033164 | 0.01 |
ENST00000409871.1
ENST00000343936.4 |
ZC3H6
|
zinc finger CCCH-type containing 6 |
| chr2_-_145277882 | 0.01 |
ENST00000465070.1
ENST00000444559.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr12_-_49351228 | 0.01 |
ENST00000541959.1
ENST00000447318.2 |
ARF3
|
ADP-ribosylation factor 3 |
| chr19_-_49314169 | 0.01 |
ENST00000597011.1
ENST00000601681.1 |
BCAT2
|
branched chain amino-acid transaminase 2, mitochondrial |
| chr17_+_39994032 | 0.01 |
ENST00000293303.4
ENST00000438813.1 |
KLHL10
|
kelch-like family member 10 |
| chr19_-_53757941 | 0.01 |
ENST00000594517.1
ENST00000601413.1 ENST00000594681.1 |
ZNF677
|
zinc finger protein 677 |
| chr14_+_21492331 | 0.01 |
ENST00000533984.1
ENST00000532213.2 |
AL161668.5
|
AL161668.5 |
| chr3_+_155838337 | 0.01 |
ENST00000490337.1
ENST00000389636.5 |
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr16_-_55866997 | 0.01 |
ENST00000360526.3
ENST00000361503.4 |
CES1
|
carboxylesterase 1 |
| chr11_-_65624415 | 0.01 |
ENST00000524553.1
ENST00000527344.1 |
CFL1
|
cofilin 1 (non-muscle) |
| chr1_+_153963227 | 0.01 |
ENST00000368567.4
ENST00000392558.4 |
RPS27
|
ribosomal protein S27 |
| chr12_-_49351148 | 0.00 |
ENST00000398092.4
ENST00000539611.1 |
RP11-302B13.5
ARF3
|
ADP-ribosylation factor 3 ADP-ribosylation factor 3 |
| chr7_-_140714739 | 0.00 |
ENST00000467334.1
ENST00000324787.5 |
MRPS33
|
mitochondrial ribosomal protein S33 |
| chr2_-_208030647 | 0.00 |
ENST00000309446.6
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr1_-_42384343 | 0.00 |
ENST00000372584.1
|
HIVEP3
|
human immunodeficiency virus type I enhancer binding protein 3 |
| chr1_+_109102652 | 0.00 |
ENST00000370035.3
ENST00000405454.1 |
FAM102B
|
family with sequence similarity 102, member B |
| chr17_+_75450075 | 0.00 |
ENST00000592951.1
|
SEPT9
|
septin 9 |
| chr5_+_32711419 | 0.00 |
ENST00000265074.8
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
Network of associatons between targets according to the STRING database.
First level regulatory network of PRDM14
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0015068 | amidinotransferase activity(GO:0015067) glycine amidinotransferase activity(GO:0015068) |
| 0.0 | 0.0 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.0 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.0 | 0.0 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.0 | 0.0 | GO:0000773 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |